The human La protein was discovered as the protein component of a heterogeneous group of small RNA-containing particles to which patients with autoimmune disorders such as Sjögren’s syndrome and systemic lupus erythematosus produce pathogenic antibodies. Molecular biologists used these antibodies as tools to isolate La, clone its gene, characterize its associated RNAs, and study their functional relationships in normal and other cells. The small RNAs associated with La are a large variety of newly synthesized precursors of the tRNAs and other transcripts produced by RNA polymerase III. We now know that a principal function of La is to bind to the common Uridylate-rich terminal motif shared by these RNAs and protect them from untimely digestion by degradative and other 3′ riboexonucleases, retaining them in the nucleus while they undergo processing and chemical modifications necessary for proper folding and maturation before export to the cytoplasm.
La protein appears to have arisen as early in evolution as RNA polymerase III itself; homologous sequences are found in the most ancient eukaryote representatives known and in all species examined it has the same principal function. Key to La’s binding and protection of RNA 3′ terminal ends is the unique structural organization of its two distinct RNA binding domains, the La Motif (LaM) and RNA recognition motif-1 (RRM1) which together are referred to as the “La Module.” Very early in eukaryotic evolution, prior to the emergence of the most primitive multicellular animal cells, La acquired a second RRM, separate from the La module, that is intimately followed by an alpha-helix, referred to as RRM2a. Recent studies revealed La as one of the ancestral repertoire of 25 RNA-binding proteins essential to animal stem cells. La also interacts and functions with certain mRNAs, of cellular and viral origin, and is engaged by the short leader RNAs of some viruses.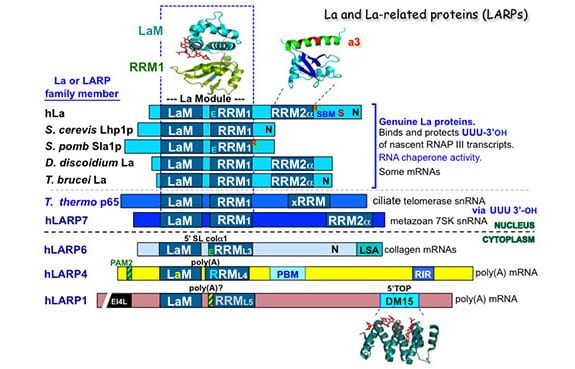
Four additional La-related proteins (LARPs) all bearing the core La module emerged in eukaryotes and established the major conserved families known as LARPs 7, 6, 4 and 1, each with unique domains following their La modules. While La was discovered more than 35 years ago, the LARPs were realized to exist about 10 years ago and the rate of their research has been accelerating swiftly. We now know that the LARPs contribute important functions in fundamental aspects of RNA metabolism. Detailed information on function of at least one member of each LARP family reveal that that RNA metabolism is central to their functions. LARP7 functions with small nuclear RNAs similar to La, whereas LARPs 6, 4 and 1 are cytoplasmic and function with mRNAs.
In a review article in WIREs RNA, we provide overviews and updates of the known structures of the La modules of the La proteins and LARPs that have been determined. We also compare and contrast their mechanisms of interactions with RNA and consider related outstanding questions. The review also provides some historical perspective on La and the LARPs as well as recent advances toward understanding their functions in the areas of mRNA stability, protein production, ribosome biogenesis and regulation thereof and for some additional roles including unexpected ones and in cancer.
Three-dimensional structures of La and LARP7 family members bound to their RNA ligands reveal that for these, the LaM and RRM form a V-like clamp around the RNA, sequestering the 3′ end at the end of a hydrophobic pocket, accounting for their 3′ protection activity. We also review a great deal of what is known from published studies of the biochemistry of RNA binding to La and different LARPs. Structures of individual domains of some of the LARPs have also been solved, some in the presence of RNA, which are also reviewed. One example is the 3D structure of the DM15 domain of LARP1 which binds to the 5’cap-TOP (terminal oligopyrimidine) motif found on the mRNAs that encode ribosome subunit proteins and helps coordinate their regulation.
Kindly contributed by the Authors.