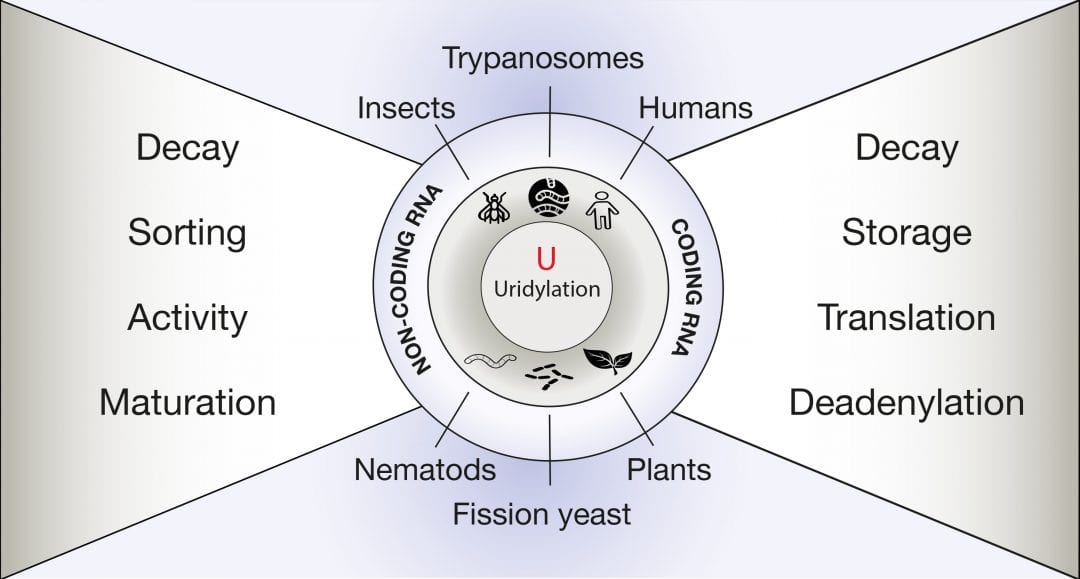
Every genome encodes thousands of coding and non-coding RNAs. Altogether, these RNAs constitute the transcriptome, whose composition is constantly regulated to optimize the cells’ responses to complex and ever-changing internal and external signals. This regulation is achieved by a plethora of post-transcriptional processes that, together with transcription, dynamically shape the transcriptome. The post-transcriptional addition of a few uridines at the 3’ end of a transcript is called RNA uridylation and is emerging as a decisive process regulating the transcriptome of a wide range of eukaryotes, from trypanosomes to plants and animals.
An advanced review in WIREs RNA by De Almeida et al. discusses the various functions that are associated with RNA uridylation, exposing why this modification must be considered as a generic feature in the metabolism of eukaryotic RNAs. The diversity of consequences linked to RNA uridylation is illustrated for both non-coding RNAs and mRNAs. RNA uridylation can control the processing, influence the activity, or trigger the elimination of various types of non-coding RNAs. In addition, messenger RNAs are also massively targeted by uridylation. The prime function for mRNA uridylation is to facilitate degradation. Yet, recent studies suggest that uridylation has additional roles, such as preventing the excessive deadenylation of mRNAs or regulating translation.
Exciting times lie ahead in this emerging field of RNA uridylation. New breakthroughs are expected to further elucidate the molecular mechanisms underlying uridylation-mediated regulation of gene expression. The full impact of RNA U-tailing during developmental transitions or in response to pathogen attacks and diseases also remains to be fully appreciated.
Kindly contributed by the Authors.