Image credit: Louis Reed on Unsplash
Cells respond to stimuli from their environment and these responses are vital for proper cellular behavior and function. In a biological setting, most mammalian cells are attached to a surface, such as the gut lining, and are guided by interactions with that surface through external stimuli — this includes surface chemistry, topography, and substrate stiffness.
In recent years, researchers have experimented with altering surface micro- and nanotopography of cells as a way to regulate their attachment and function. However, despite advancements in this area, there is no effective way to screen surface geometries to identify the exact cues that influence cell function. Add to this the huge variety of surface geometries in naturally occurring cell culture substrates, which makes it impractical for scientists to replicate and test them all in a lab.
But what if researchers could build a predictive model by finding a way to screen a select number of structures?
Scientific efforts have turned to the high-throughput screening potential of topographical platforms, fabricated through a range of methods and materials.
Unfortunately, the different materials used can act as an uncontrolled variable, making it extremely difficult to ascertain whether results are comparable across studies. In addition, it remains a challenge to translate these findings to fundamental biological research in which cells are typically grown on conventional tissue culture plastic (TCP) — a transparent polystyrene that is treated with plasma to mimic natural structures and provide a suitable platform for cell adhesion.
How can we do it better?
A research team led by Dr Victor Cadarso of Monash University, Australia, determined that imprinting surface topographies — those that could influence the function of cells in the well — directly onto a TCP plate would enable direct comparison between screening experiments and fundamental research.
They developed a method to create the customizable micro- and nanostructured screening motifs using 3D printing, through a technique called thermal nanoimprint lithography (NIL), which is scalable and well-developed. These motifs were imprinted on the base of standard TCP multi-well plates to make the method compatible with high-throughput screening.
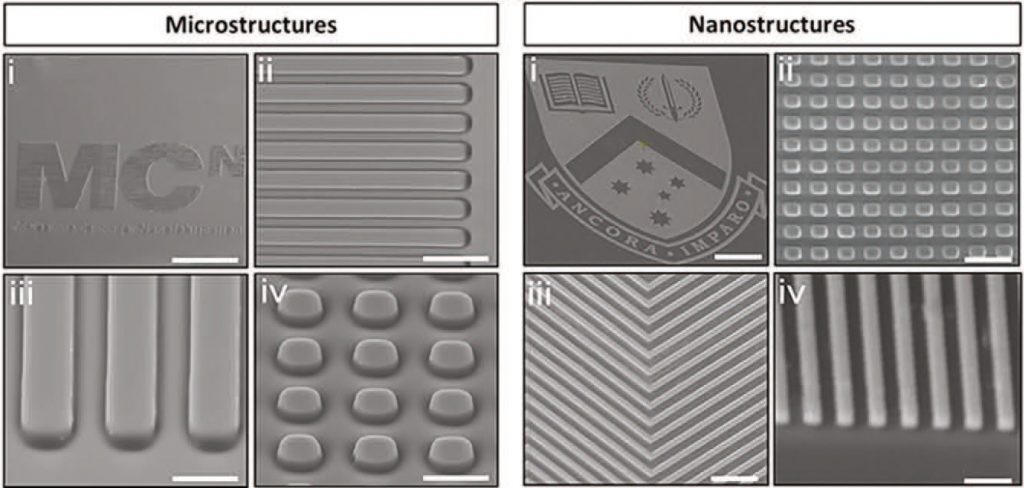
Crucially, there was no change to the chemical, mechanical, or optical properties of the material, thus eliminating the uncontrolled variables that hindered previous methodologies.
To test their new system, the team applied their plates to screening across five distinct cell types. They assessed the effects of culture material on cell morphology and function.
Cells were cultured on conventional TCP multi-well plates, TCP plates imprinted with a flat motif (to mimic the conventional plate), and Ormocomp (a polymer commonly used for microfabrication) substrates. There were negligible differences between the TCP-plate cultures, but noticeable changes in the Ormocomp-culture. This confirms that imprinting does not affect cell behavior and may be more suitable than existing technologies.
The team then screened different topographies, noting how differences in the structure directly influenced cell function and behavior.
This new method of imprinting TCP plates can now be used in a wide range of investigations looking into the effects of microtopography on cell morphology.
The simple technique can be combined with existing protocols and high-throughput screening. In addition, it is easy to scale up — even for large-scale manufacturing procedures. The team hope that this cost-efficient and standardized method will advance mainstream biological work including drug discovery, tissue engineering, and cell therapy.
Reference: J. Carthew, et al., Next Generation Cell Culture Tools Featuring Micro- and Nanotopographies for Biological Screening, Advanced Functional Materials (2021). DOI: 10.1002/adfm.202100881