While we rarely think of the polymers of life (proteins, polynucleotides and polysaccharides) in the same way as synthetic polymers, it is sometimes interesting to do so. There is a theory in the chemical origins of life that holds that polynucleotides (RNA, DNA) predate proteins; that there was, in fact, an “RNA world”. But RNA replication is facilitated by enzyme (protein) catalysts. Is accurate replication of RNA possible by nonenzymatic means?
While we rarely think of the polymers of life (proteins, polynucleotides and polysaccharides) in the same way as synthetic polymers, it is sometimes interesting to do so. There is a theory in the chemical origins of life that holds that polynucleotides (RNA, DNA) predate proteins; that there was, in fact, an “RNA world”. But RNA replication is facilitated by enzyme (protein) catalysts. Is accurate replication of RNA possible by nonenzymatic means?
The critical test of these ideas is the number of correct nucleic acid additions that is likely before an incorrect addition (error threshold). Rajamani et al. in 2010 invoked the concept of stalling to extend the error threshold to values thought to be viable for chemical evolution. The concept of stalling postulates that, in the event of an incorrect nucleic acid addition in the penultimate position, all additions (correct and incorrect) in the ultimate position are suppressed – presumably by steric effects.
Thus, RNA strands with errors will replicate more slowly than those without, and so will not complete effectively for the available nucleic acids. In addition to some very elegant kinetic measurements of monomer addition rates, Rajamani has modified the calculation of the error threshold to account for stalling.
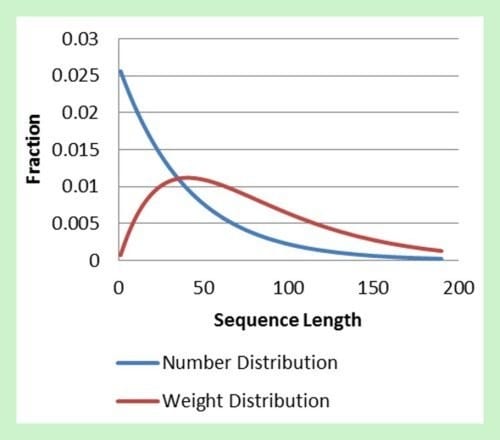
Joe Schork at the Georgia Institute of Technology in Atlanta now has shown that Rajamani’s mechanism of stalling can be accounted for by using the classical Mayo-Lewis Equation for calculating copolymer sequence lengths in copolymerizations of commercial synthetic polymers. Using the kinetic parameters of Rajamani it is possible to independently predict the error threshold for RNA replication. Agreement with the model of Rajamani is surprisingly good.