Biological systems are complex, meaning that they exhibit nonlinear behaviors due to positive and negative feedback, as well as dependence on the biological and temporal context in which they take place. The inflammatory response, which is needed to appropriately resolve injury, infection, and stress–and the root of many diseases–is a prototypical complex system. While such complex systems often defy reductionist analysis, mechanistic computational modeling has been used to describe such systems and make non-intuitive predictions.
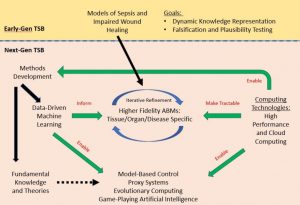
Agent‐based models (ABMs) of inflammation: past, present, and future.
Agent-based modeling is one of several mechanistic computational modeling approaches used in various disciplines. Compared to the more typical modeling frameworks involving differential equations, agent-based modeling offers several key advantages. Agent-based models are a form of “bottom-up” modeling, which means that models are based on individual actors (e.g. molecules, cells, people) and the emergent phenomena that occur when they interact with each other is based on rules that are applied in a stochastic fashion. Thus, agent-based models are relatively easy for non-specialists to understand. Although, these models can require extensive computational resources and can be more difficult to parameterize compared to equation-based models.
A decade ago, Yoram Vodovotz and Gary An summarized the then-nascent state of agent-based modeling as applied to the inflammatory response, with an explicit focus on clinical applications. They called the overall approach, “Translational Systems Biology.” In the intervening years, many methodological advances have occurred, and multiple research groups have adopted this methodology in diverse studies that involve inflammation. This WIREs Systems Biology and Medicine review describes these developments over the past decade, detailing some of the remaining challenges and the widespread use of this approach, while also suggests ways by which these challenges could be overcome.