Over the past few years, artificial intelligence (AI) has emerged in the drug discovery field. AI drug design is developed from the simplified molecular-input line-entry system (SMILES) string to a graph neural network which has improved accuracy and efficiency of de novo drug design discovery. AI is not only able to design drugs based on the trained deep learning model, but can also employ deep learning models for predictions of retrosynthesis and drug properties such as ADMET (absorption, distribution, metabolism, excretion, and toxicity). AI is considered to be a promising way to find more compound candidates and raise the success rate of these candidate’s clinical trials.
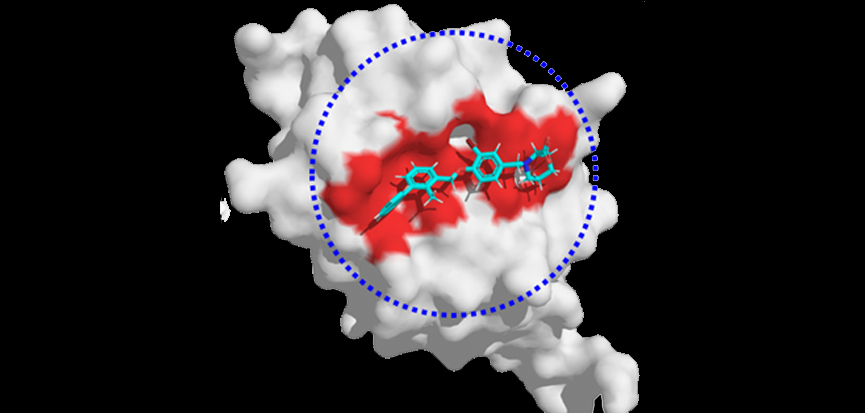
Prof. Qifeng Bai who works at Lanzhou University in China thinks AI will evolve de novo 3D drug design from 1D or 2D drug design. Bai’s group has developed a software named MolAICal which can design 3D ligands in the pocket of disease targets by AI and classical algorithms. With the development of AI technology, AI has been developed for drug molecular docking. For instance, GNINA is an excellent tool based on convolutional neural networks (CNNs) — neural networks primarily used for analyzing visual imagery — for molecular docking. Moreover, molecular dynamics (MD) simulation is a dynamical means that can improve the precision of drug design. Even though MD simulation is considered a more accurate way than stationary de novo drug design, it costs time and computing resources to deal with high dimensional structural space of large complex simulated systems. AI can provide an effective and economical way to deal with the big data of MD simulations, and play an important role in stationary and dynamical de novo 3D drug design for our benefit.
Dr. Horacio Pérez-Sánchez who leads the Structural Bioinformatics and High Performance Computing (BIO-HPC) research group at UCAM Universidad Católica de Murcia thinks that even if we realize the outstanding results obtained in the last decade with neural networks and other machine learning architectures, we should also understand why those predictions are made and how. That is the main objective of Interpretable Machine Learning (IML) methods that have been gaining popularity in the last five years to provide such information and rules about the internal workings and logic of such “black boxes” and which can help tremendously in all drug discovery fields.
AlphaFold2 has proved to be of great value for AI, biology and medicine. It inspires academics and industry to develop more AI tools for drug discovery, reducing cost and time to the market. Various directions have made progress, including an AI-based scaffold hopping method to jump out of patent protection for novel drugs, protein structure prediction, virtual screening, generative chemistry, ADMET prediction, and synthetic route planning based on deep learning models. The problems faced in drug discovery also lead the development of deep learning such as deep Graph Learning, physics-driven deep learning, energy-based models, and so on.
Generally speaking, AI has been gradually introduced into every stage of drug discovery including preclinical studies and clinical trials Phases I, II, III, IV, and so on. We believe that AI will accelerate the speed of drug development and help humankind overcome more diseases.
Written by: Qifeng Bai, Tingyang Xu, and Horacio Pérez-Sánchez
Reference: Q. Bai et al., ‘Application advances of deep learning methods for de novo drug design and molecular dynamics simulation‘, WIREs Computational Molecular Science (2021) DOI: 10.1002/wcms.1581