Image credit: Patrick Seeger/University of Freiburg
Antibiotic-resistant bacteria are on the rise with new strains capable of evading front-line medications springing up all over the world. While researchers scramble to identify new classes of drugs to bridge this gap, bottlenecks in this process mean we are falling behind and are approaching an era where even a simple infection could once again kill.
Researchers at the University of Freiburg led by Dr. Can Dincer and Professor Wilfried Weber think that one strategy to slow this decline is to optimize the concentrations of antibiotics we still have to avoid the emergence of drug resistance due to insufficient dosing.
“This problem mandates the re-evaluation of our battle strategy about how we utilize the existing antibiotics,” explained Dincer in an email to Advanced Science News. “Recent clinical data reveal that the development of the antimicrobial resistance could be slowed down by optimizing the antibiotic dosage regimen.”
Currently, clinicians lack the ability to effectively monitor the concentration of antibiotics in patients, which would require frequent blood tests and monitoring. This can be problematic as the success of a therapy depends on antibiotic concentrations remaining within a specific therapeutic window — this is unique depending on how a patient’s body responds and breaks down the medication. Operational windows have instead been determined based on data collected from animal models and healthy volunteers, resulting instead in ranges of statistically acceptable concentrations: subtherapeutic, therapeutic, or toxic.
“It means, most of the time, patients cannot receive the optimal doses for their current condition,” said H. Ceren Ates, a researcher at Freiburg and the study’s first author. “With the motto ‘nobody is average’ in mind, we would like to respond this need.”
Dincer and Ates think that this could be achieved by administrating the correct dose at the right time through tailored therapies. Currently, if a course of antibiotics fails to completely eradicate an infection, then additional therapeutics are used or added to the course of treatment. Instead of a “one-size-fits all” approach, the team sought to develop an easy and cost-effective means of monitoring antibiotic levels for a patient’s individual needs.
“We need to find out how a drug is metabolized in the body considering several parameters like individual pharmacokinetics, age, or comorbidity,” said Dincer. “[This] personalized scheme, however, requires frequent sampling and analysis.” While this would help curb the ineffective use of antibiotics, this workflow is not feasible for most healthcare providers to manage for every patient.
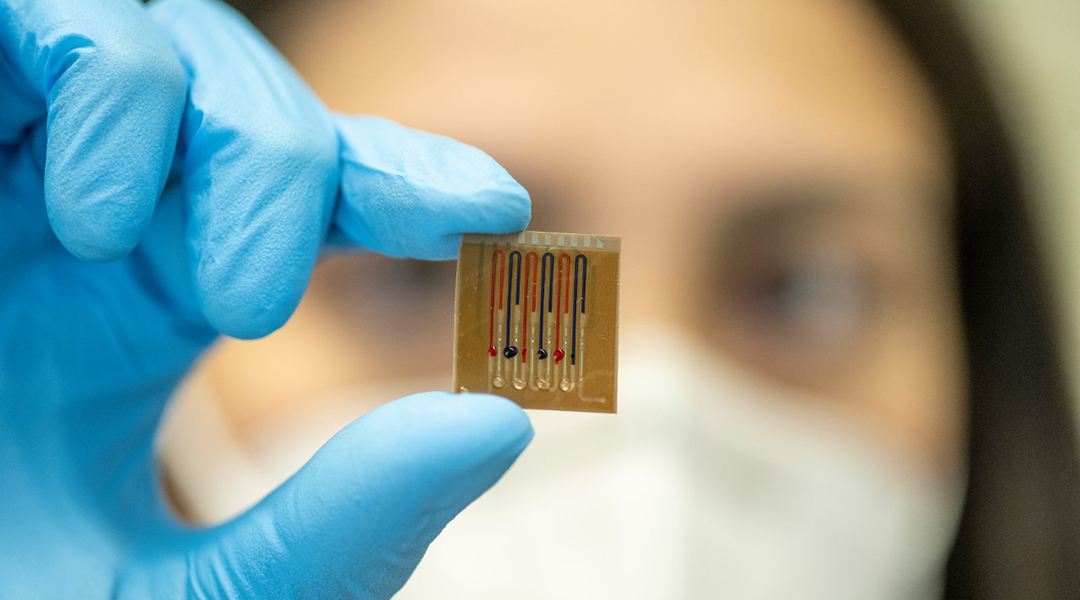
To overcome this roadblock, Dincer and his team have developed a disposable, antibody-free microfluidic biosensor capable of quickly and accurately monitoring up to four drugs (with the possibility to extending this to eight) in the body on the same chip. It incorporates immobilized biomolecules on a surface within the device, which elicit a change in an electrochemical signal when they come into contact with specific molecules, such as a designated drug molecule.
“One of the unique features of our sensing technology, which is different from most of its counterparts, is that the section for biomolecule immobilization is separated from the electrochemical detection area,” said Ates. “With such a configuration, we inherently bypass the electrode [contamination] issue and can operate complex biofluids like untreated, whole blood without compromising sensitivity.”
The high sensitivity of the device allowed the team to identify even trace amounts of drug levels in exhaled breath, producing an antibiotic “breathalyzer” of sorts. “We were able to find a correlation between measured [exhaled breath condensate] and blood levels, for the first time,” said Ates.
The device is boasted as being “antibody free”, a common protein-based detection strategy. There are two main advantages to this, says Dincer. The first is increased sensor stability, as antibodies are temperature sensitive, limiting how they can be stored and for how long. Secondly, and perhaps the most important advantage, antibodies make this device quite expensive. “Using antibodies increases the production cost of a single chip at least 10-fold, which contradicts our first objective of developing an easy-to-use, low-cost technology for frequent measurement of antibiotic levels for better drug management,” said Ates.
The device’s capabilities were demonstrated in animal models, accurately predicting different β-lactam concentrations in different biofluids, such as blood and saliva. The team is now working to miniaturize their system to make it more user friendly, as well as improve sample-to-result time with the ultimate goal of providing results in under 30 minutes — the device currently requires 90 minutes.
“[We hope] to integrate our system in a real-life therapeutic drug monitoring scenario next, where we would like to observe the effect of biosensor-based dose adjustment recommendations on the clinical response,” added Dincer. “After this study, we believe that our system will be ready to be used in the clinic as a supplementary tool.”
Reference: H. Ceren Ateş , et al, Biosensor-Enabled Multiplexed On-Site Therapeutic Drug Monitoring of Antibiotics, Advanced Materials (2021). DOI: 10.1002/adma.202104555