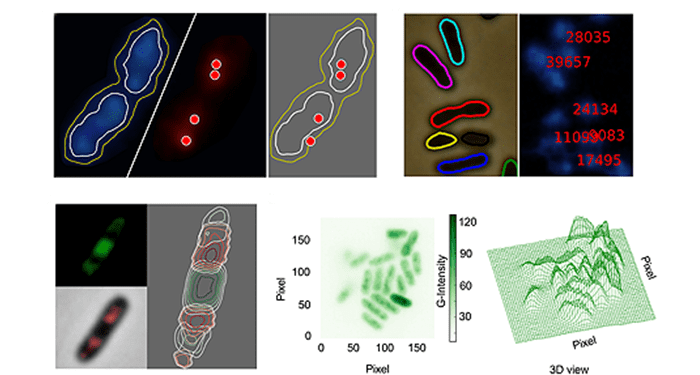
For scientists who study the metabolic engineering of bacterial cells, visualization of the intracellular constituents of individual bacteria while they act as live biocatalysts is emerging as a key research area. Thus the rigorous analysis of fluorescence microscopy images is of critical importance. Unfortunately, the currently available bioinformatic tools generally lack flexibility and are often subject to licensing or other limitations. In this context, Goñi-Moreno and colleagues have developed CellShape, a user-friendly image analysis tool for quantitative visualization of bacterial cell factories, which is easy to use even for non-experts.
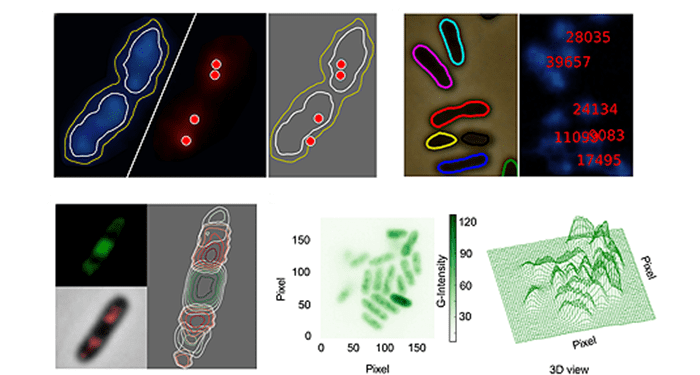
CellShape is coded in Python, a free, open-source programming language with widespread community support. CellShape uses subpixel clusters or contours as the fundamental methodology, which can be fine-tuned by the users. In order to prove the value of this platform, Goñi-Moreno and colleagues adopted this platform to analyze the tridimensional distribution of the constituents of the gene expression flow (DNA, RNA polymerase, mRNA and ribosomal proteins) in an individual industrial strain Pseudomonas putida KT2440.
In the present CellShape version (v0.8), the authors have left a considerable room for improvement in the future, and this platform can be applied to virtually any type of bacteria analyzed with fluorescence microscopy. With the growing interest un understanding the intracellular interactions of bacterial organelles, fluorescence labelling and reading technologies are becoming more and more important in the industrial biotechnology research field. Therefore, CellShape, the free and open-source platform, is promising and appealing for the scientific community.