Single nucleotide variations (SNVs) are types of mutations in the DNA sequence affecting the ways different people respond to diseases and treatments. People inherit some SNVs from their parents, while they acquire others—such as oncogenic ones—over the course of their lives. Scientists have been able to detect extensive arrays of SNVs in human diseases due to the development of next-generation sequencing technologies.
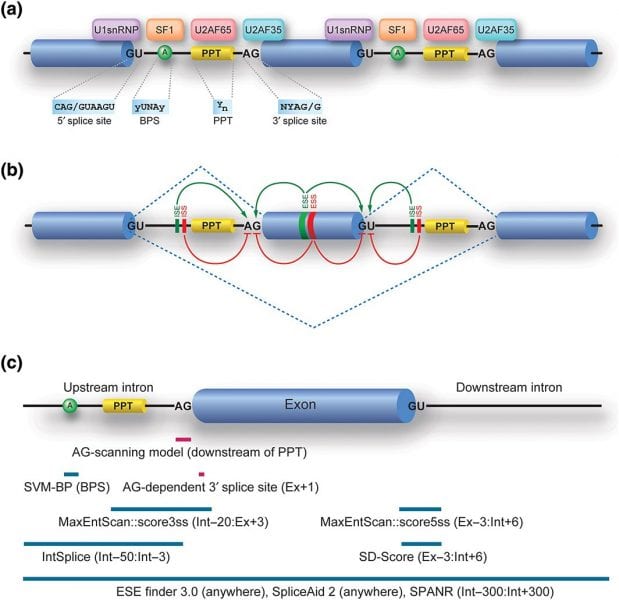
Alternative RNA splicing is the most essential and most studied of the RNA processing mechanisms used to achieve proteome diversity. More than 90 percent of genes are alternatively spliced in a process where intronic nucleotide sequences are removed and the remaining exonic sequences join together. The effects of splicing of exonic and intronic SNVs can be experimentally proven, but researchers cannot individually analyze the large number of SNVs identified in individual patients. While there are a variety of tools to predict the splicing effects of SNVs, determining the splicing effects of intronic SNVs located away from exons remains very challenging.
In the review article “Rules and tools to predict the splicing effects of exonic and intronic mutations” recently published in WIREs RNA, Kinji Ohno, Jun-ichi Takeda, and Akio Masuda of Nagoya University Graduate School of Medicine examine the current tools available to predict RNA splicing effects. In their review, the authors search for the genetic causes of human disease by identifying a large number of SNVs. Most exonic SNVs introduce missense/nonsense codons and most intronic SNVs are silent, but some affect auxiliary splicing cis-elements or generate cryptic GT-AG dinucleotides. Only two tools, the Human Splicing Finder (HSF) and the SVM-BP finder, are available to predict the position of the branch point sequence. Similarly, IntSplice and SPANR are the only tools to predict the splicing effects of intronic SNVs. Unfortunately, none of these can yet ensure 100% accuracy. In the future, the authors hope geneticists will be able to identify hidden splicing rules and develop further efficient tools to facilitate the understanding of splicing pathomechanisms in human diseases.
Kindly contributed by WIREs editorial staff and the authors.