Metastases are formed in a stage of cancer progression marked by the dissemination of neoplastic cells from the tumor’s site of origin to distant organs. If the disease achieves this phase, it is generally considered incurable. Among the biological processes driving metastasization, the epithelial-to-mesenchymal transition (EMT) plays a fundamental role.
EMT is involved in numerous physiological processes, such as embryonic development, but can be hijacked by cancer to provide cells with the ability to leave their site of origin and enter the bloodstream. In this respect, preventing or safely redirecting EMT in cancer has been the main goal of a number of research projects.
Unfortunately, no effective treatment strategies based on this concept are available at the present time. This is partly due to the complexity of EMT and its ability to adapt to different environments and cell types.
Computational models have recently been used to tackle this challenge, providing researchers with new tools to study EMT, understand its triggering and evolution, and potentially drive it through carefully crafted stimuli. This integrated experimental/computational approach is discussed in a recent review published in WIREs Systems Biology and Medicine and authored by a multidisciplinary team of engineers, medical doctors, and cellular and molecular biologists.
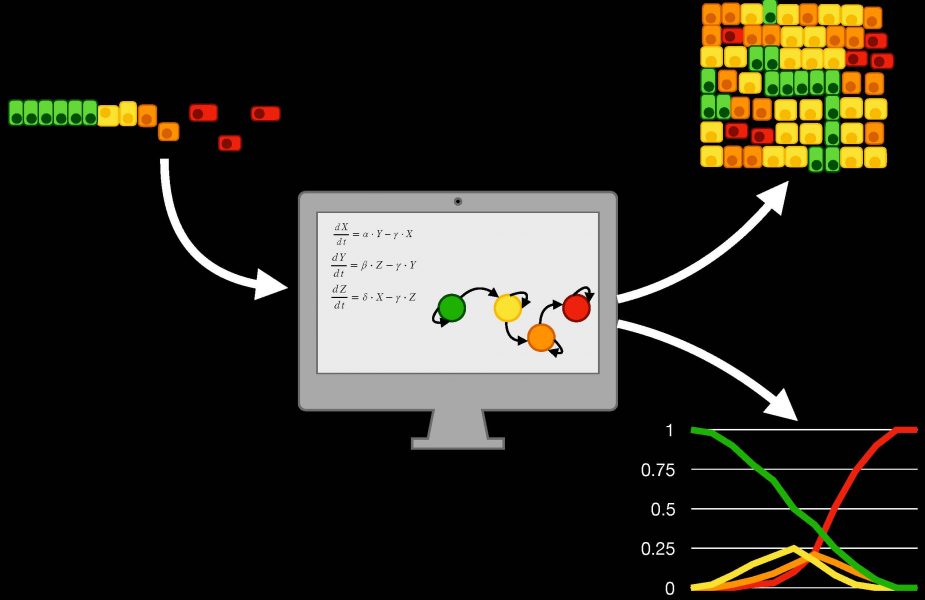
The team collected and closely analyzed mathematical representations of the changes in gene expression induced by EMT, showing how these tools can provide insight into the role of specific proteins involved in this transformation, or simulate the behavior of a population of cancer cells and their interactions with the environment.
“It is an entirely new way of conducting experiments,” said Dr. Cortesi, first author of the paper, “that allows you to easily test the effect of switching on and off different components, changing experimental conditions, length and temporal resolution of the analysis.”
This approach is particularly relevant for the study of EMT and other complex biological processes that are the result of the coordinated activity of multiple signal transduction pathways. Indeed, computational models can process large amounts of information and well-established techniques allow the extraction of relevant features, such as which genes better recapitulate EMT and the role of specific markers within this transition. Aside from providing useful mechanistic insights on cancer progression and metastases formation, this knowledge can aid in the identification of novel therapeutic targets and treatment strategies.
Recent advances in EMT computational modelling couple the representation of gene expression changes with formalisms that take into account spatial variables, such as the position of the cells and the characteristics of the microenvironment. These frameworks, called multiscale models, effectively capture EMT complexity at different resolutions (i.e., at the single cell and at the population level), allowing researchers to study how changes occurring in single cells propagate through the population and how spatial patterns emerging in a group of cells undergoing EMT affect the behavior of its components.
“Multidisciplinary research has the fundamental edge of combining different expertises and knowledge to achieve a common goal,” remarked Dr. Cortesi, “and, as such, it has the potential of fostering truly innovative solutions, improve our understanding of complex biological phenomena and aid the development of effective new treatments.”
Reference: Marilisa Cortesi et al. ‘Computational models to explore the complexity of the epithelial to mesenchymal transition in cancer.’ WIREs Systems Biology and Medicine (2020). DOI: 10.1002/wsbm.1488