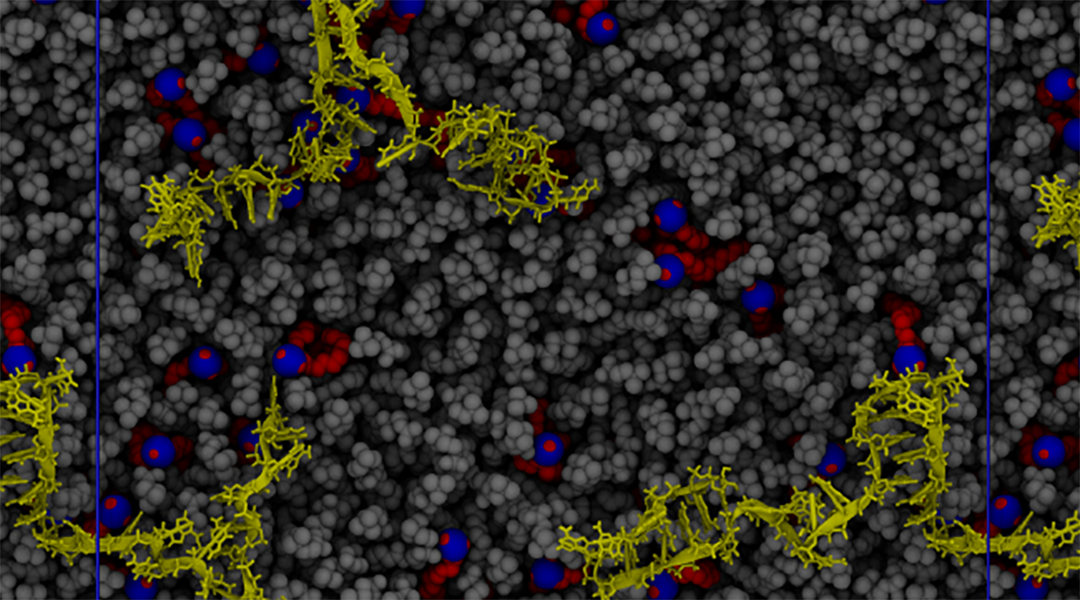
Image credit: G. Settani et al., Macromol. Rapid Commun. 2021, 2100683
Thanks to companies like BioNTech, Pfizer, and Moderna, mRNA vaccines have become a buzzword in 2021. One of the keys to their success is the means by which mRNA is transported to our immune cells: lipids.
Lipid-based materials such as lipoplexes, lipid-based nanoparticles (LNPs), and liposomes, are known to be a fundamental tool for drug — and mRNA — delivery. Last year, researchers highlighted this again using liposomes to encapsulate SARS-CoV-2 antibodies.
For vaccines, lipid-based delivery systems generally exhibit less side effects in patients compared with traditional viral vectors. The whole delivery process of these mRNA molecules — as well as other drugs — is called transfection, and whilst lipid-based delivery systems are better than viral vectors in some ways, their transfection efficiency is generally lower.
Researchers have been able to narrow down their choice of lipids to improve transfection efficiency, and nowadays, formulations are usually a mix of an ionizable cationic lipid — which is positively charged and is therefore well-suited for binding to negatively charged mRNA — cholesterol, a helper lipid, and a polyethylene glycolated lipid. However, an understanding of the exact chemical processes taking place in these lipid–mRNA interfaces is lacking. But a team of researchers have turned to computer models to better understand the chemistry between lipid-based transfection.
Friederike Schmid and Giovanni Settanni at the Johannes-Gutenberg University, Mainz, teamed up with Wolfgang Brill and Heinrich Haas at BioNTech to systematically study these interactions as a function of pH in order to hopefully guide experimentalists on their lipid-of-choice when optimizing transfection efficiences of their mRNA vaccines. With other nucleic acid-based therapeutics gaining traction, such as gene therapy and siRNA therapy, understanding the fundamentals of transfection efficiency in this area is of growing importance.
Schmid and her colleagues looked to the lipid 2‐dioleyloxy‐N,N‐dimethyl‐3‐aminopropane, or DODMA, where experiments have shown that DODMA and mRNA form a structure wherein a lipid bilayer of DODMA is separated by a layer of mRNA, with a distance between the layers ranging from about 6.5 to 8 nanometers.
Exactly how RNA is encapsulated in this DODMA-based system determines both the level of protection against degradation from the host organism, and the way RNA will escape the endosome — that is, the structures between cells that “sort” material entering them — after the fusion of the nanoparticle with the endosomal membrane.
The computer models the researchers used were molecular dynamics simulations; a powerful tool that has already been used to help drug design, and last year helped elucidate the structure of SARS-CoV-2 at the molecular level.
They found that at low pH, there is more attractive force between DODMA and mRNA, and at intermediate to high pH, these forces become repulsive. This change in pH conditions and the electrostatic interactions between the lipid and RNA affects where the RNA is found within the bilayer structure. At low pH, the RNA tends to clump around the lipid headgroups, while at higher pH, the RNA is more evenly distributed within the lamellar structure, which is hydrophilic. Furthermore, the attractive forces at low pH stretch the RNA molecule, whereas at higher pHs this biopolymer is less stretched and more confined.
The authors hope that their results can help researchers tailor future lipid-based mRNA delivery systems by fine-tuning the chemistry of the lipids used, so as to improve mRNA circulation, cellular uptake, and endosomal processing.
Reference: Geovanni Settani et al., pH‐Dependent Behavior of Ionizable Cationic Lipids in mRNA‐Carrying Lipoplexes Investigated by Molecular Dynamics Simulations, Macromolecular Rapid Communications (2021). DOI: 10.1002/marc.202100683