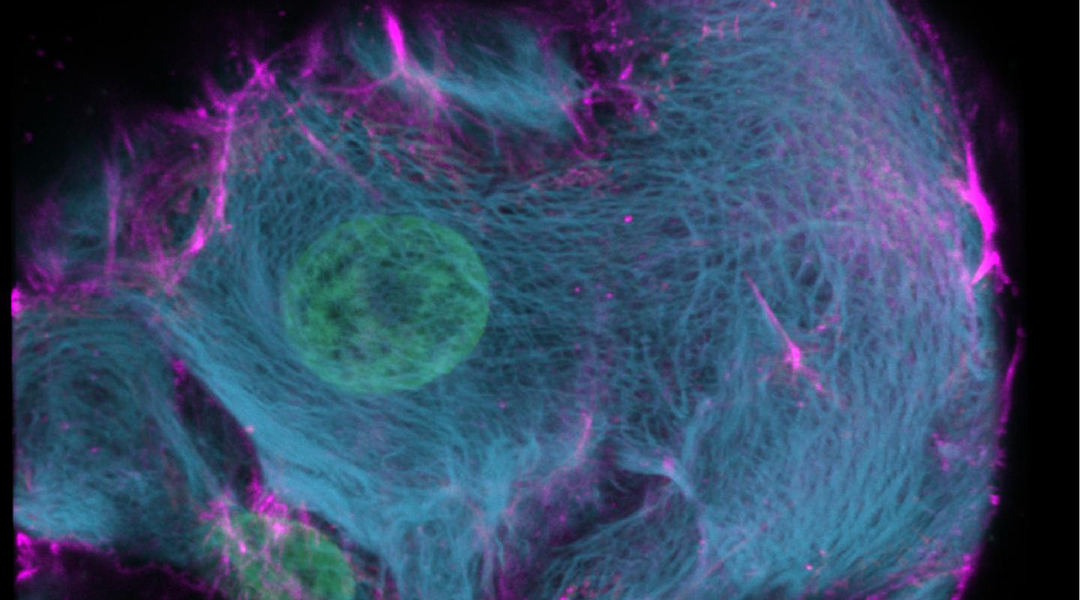
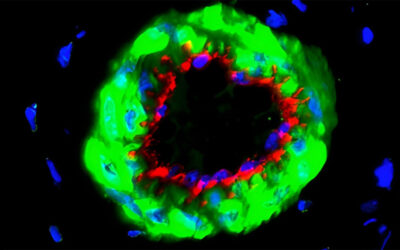
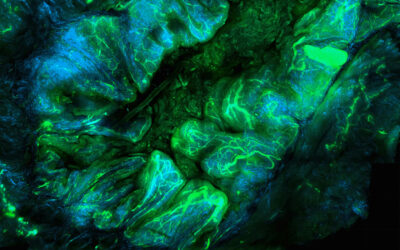
Researchers from the Hubrecht Institute, the Netherlands, have reported a variant of the CRISPR enzyme that can label specific genes in models of human organs called organoids. In the study, the new enzyme, called CRISPR-HOT, was used to visualize individual cell types and sub-cellular components, such as protiens, using gene editing to incorporate different fluorescent tags to ultimately investigate how the deletion of a specific gene in liver cells (TP53) may lead to cancer as a result of uncontrolled cell division.
In the last decade, CRISPR/Cas9 has revolutionized gene editing. The original enzyme is a component of bacterial immune systems, where its purpose is to cut and break the DNA of invading viruses. In the last decade, the hype around CRISPR-based technologies has grown with countless innovative applications having been reported, from engineering DNA nanostructures to treating debilitating genetic diseases such as sickle-cell anemia.
“CRISPR/Cas9 can be used in many ways for different genome editing purposes,” said Benedetta Artegiani, a Ph.D. student at the Hubrecht Institute and one of the study’s authors. “Classical CRISPR/Cas9 approaches generate cuts in the genome with the purpose of introducing small mutations or inserting big portions of DNA by exploiting one of the mechanisms used by the cells to repair these breaks. However, this last process is extremely inefficient. CRISPR-HOT solved this low efficiency limitation by instead using the other mechanism of DNA repair that was previously thought not to be very feasible.”
This repair mechanism is referred to as non-homologous end joining, and was used in early studies to insert new DNA into mice. Here, the cell’s machinery attempts to repair the cut by “pasting” the DNA strand back together. “Using this machinery, we can instead also provide the cells with a piece of foreign DNA that is then used, sort of as a band aid, to close the wound,” continued Artegiani. “By this approach, we can at a specific spot in the DNA introduce the foreign DNA that can then for example be used to visualize genes by making fluorescently labelled proteins.”
And while not a widely used method, the Hubrecht team led by Professor Hans Clevers set out to test this in human organoids, finding it to be more efficient and robust than other widely used procedures.
“CRISPR-HOT turned out to be more efficient than the original approach to make these knock-in organoids,” said Delilah Hendriks, another of the study’s authors. “In addition, the time required to make these genomic modifications is much less. The limitation of CRISPR-HOT is given by the cell itself: to visualize the gene(s), those need to be ‘activated’ by the cells, otherwise even if the fluorescent tag would be inserted this would not be visible. In addition, one limitation, not linked to the technique, but rather to the organoid model is the ability of the different cell types to allow genome engineering.”
The team successfully inserted fluorescent labels onto specific genes using their CIRSPR-HOT enzyme, creating what the group called “painted cells”, which fluoresced in different colors, allowing the team to easily identify and analyze them. They were able to do this with a class of cells in the intestine (called enteroendocrine cells), which have previously been difficult to study because they are so rare. They were also able to color organoids derived from a specific cell type in the liver to visualize individual cellular components, such as proteins, which allowed them to monitor their function and organization within the cell.
In addition, the team demonstrated a proof-of-concept in which the unstructured cell division of abnormal liver cells, which have been linked to cancer, were visualized for the first time. According to the team, adults contain higher levels of these abnormal cells, which contain more DNA than a normal cell. Clever’s team were able to label specific cellular components within the organoids that are related to cell division, and monitor how these abnormal cells divide relative to normal cells. The team was also able to use their CRISPR enzyme to easily study mutations in the TP53 gene, which has been linked to this abnormal cell division found commonly in liver cancer.
“We hope that our study contributes to answering complex questions in developmental biology and cancer biology which will be facilitated by visualizing genes through CRISPR-HOT,” stated Hendriks.
While still early days, the ease with which the human organoids could be edited, labeled, and manipulated to study different biological processes with high precision and model disease states will change the way in which we study cells, determine disease mechanisms, and perhaps develop life saving therapeutics.