Cyclic-di-GMP is a widely used nucleotide signaling molecule in bacteria. Of particular interest is how this signaling pathway regulates bacterial pathogens and their interactions with mammalian hosts. However, studying the role of cyclic-di-GMP in bacterial pathogenesis is complicated by several factors. Many bacterial genomes can contain upwards of 60 genes that encode enzymes that make or degrade cyclic-di-GMP.
Once made, cyclic-di-GMP can interact with cellular receptors including many types of proteins and RNA elements called riboswitches. Binding of cyclic-di-GMP to receptors can either activate or inactivate the function of each protein receptor or riboswitch. However, as the interaction of cyclic-di-GMP with its receptor is specific for each receptor, each cyclic-di-GMP receptor interaction must be assessed empirically.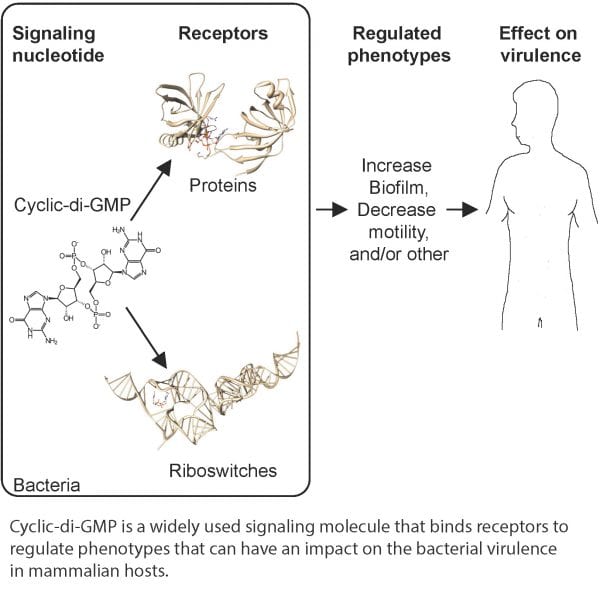
To add another degree of complication, the genome of each pathogen encodes different types of enzymes and receptors, thus making it difficult to form generalized conclusions on the ability of cyclic-di-GMP to regulate bacterial pathogenesis
In a recent review in WIREs RNA, Hall and Lee provide an overview of current literature regarding eight bacterial pathogens and how cyclic-di-GMP regulates pathogenesis.
It covers three categories of bacteria: 1) healthcare-associated pathogens, 2) gastrointestinal pathogens, and 3) vector-borne pathogens. Each pathogen was assessed for their cyclic-di-GMP regulated processes associated with mammalian infections.
For pathogens that have a reservoir outside the mammalian host, the contribution of cyclic-di-GMP signaling in those environments are also discussed. By comparing and contrasting pathogens in each category, a view emerges where each pathogen utilizes the cyclic-di-GMP pathway in different ways depending on the processes that are regulated by the signaling molecule.
Kindly contributed by the Authors.