 In the 1960s, Nobel laureate Pierre-Gilles de Gennes postulated that someday researchers could test his theories of polymer networks by observing single molecules. Researchers at Brown have now observed single molecules of DNA being drawn through nanopores by electrical current and have figured out why they most often travel head first.
In the 1960s, Nobel laureate Pierre-Gilles de Gennes postulated that someday researchers could test his theories of polymer networks by observing single molecules. Researchers at Brown have now observed single molecules of DNA being drawn through nanopores by electrical current and have figured out why they most often travel head first.
If you want to understand a novel, it helps to start from the beginning rather than trying to pick up the plot from somewhere in the middle. The same goes for analyzing a strand of DNA. The best way to make sense of it is to look at it head to tail.
Luckily, according to a new study by physicists at Brown University, DNA molecules have a convenient tendency to cooperate.
The research, published in the journal Physical Review Letters, looks at the dynamics of how DNA molecules are captured by solid-state nanopores, tiny holes that soon may help sequence DNA at lightning speed. The study found that when a DNA strand is captured and pulled through a nanopore, it’s much more likely to start the journey at one of its ends, rather than being grabbed somewhere in the middle and pulled through in a folded configuration.
“We think this is an important advance for understanding how DNA molecules interact with these nanopores,” said Derek Stein, assistant professor of physics at Brown, who performed the research with graduate student Mirna Mihovilovic and undergraduate Nick Hagerty. “If you want to do sequencing or some other analysis, you want the molecule going through the pore head to tail.”
Research into DNA sequencing with nanopores started a little over 15 years ago. The concept is fairly simple. A little hole, a few billionths of a meter across, is poked in a barrier separating two pools of salt water. An electric current is applied across the hole, which occasionally attracts a DNA molecule floating in the water. When that happens, the molecule is whipped through the pore in a fraction of a second. Scientists can then use sensors on the pore or other means to identify nucleotide bases, the building blocks of the genetic code.
The technology is advancing quickly, and the first nanopore sequencing devices are expected to be on the market very soon. But there are still basic questions about how molecules behave at the moment they’re captured and before.
“What the molecules were doing before they’re captured was a mystery and a matter of speculation,” Stein said. “And we’d like to know because if you’re trying to engineer something to control that molecule — to get it to do what you want it to do — you need to know what it’s up to.”
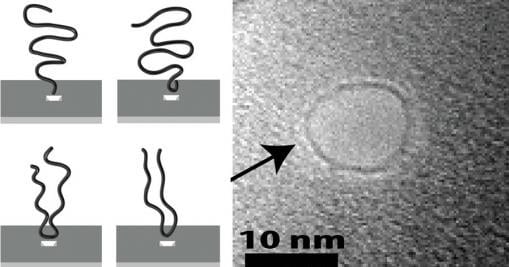
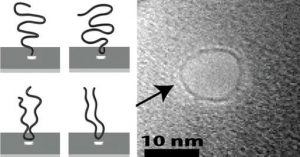
To find out what those molecules are up to, the researchers carefully tracked over 1,000 instances of a molecule zipping through a nanopore. The electric current through the pore provides a signal of how the molecule went through. Molecules that go through middle first have to be folded over in order to pass. That folded configuration takes up more space in the pore and blocks more of the current. So by looking at differences in the current, Stein and his team could count how many molecules went through head first and how many started somewhere in the middle.
The study found that molecules are several times more likely to be captured at or very near an end than at any other single point along the molecule.
“What we found was that ends are special places,” Stein said. “The middle is different from an end, and that has a consequence for the likelihood a molecule starts its journey from the end or the middle.”
Always room for Jell-O
As it turns out, there’s an old theory that that explains these new experimental results quite well. It’s the theory of Jell-O.
Jell-O is a polymer network — a mass of squiggly polymer strands that attach to each other at random junctions. The squiggly strands are the reason Jell-O is a jiggly, semi-solid. The way in which the polymer strands connect to each other is not unlike the way a DNA strand connects to a nanopore in the instant it’s captured. In water, DNA molecules are jumbled up in random squiggles much like the gelatin molecules in Jell-O.
“There’s some powerful theory that describes how many ways the polymers in Jell-O can arrange and attach themselves,” Stein said. “That turns out to be perfectly applicable to the problem of where these DNA molecules get captured by a nanopore.”
When applied to DNA, the Jell-O theory predicts that if you were to count up all the possible configurations of a DNA strand at the moment of capture, you would find that there are more configurations in which it is captured by its end, compared to other points along the strand. It’s a bit like the odds of getting a pair in poker compared to the odds of getting three of a kind. You’re more likely to get a pair simply because there are more pairs in the deck than there are triples.
This measure of all the possible configurations — a measure of what physicists refer to as the molecule’s entropy — is all that’s needed to explain why DNA tends to go head first. Some scientists had speculated that perhaps strands would be less likely to go through by the middle because folding them in half would require extra energy. But that folding energy appears not to matter at all. As Stein puts it, “The number of ways that a molecule can find itself with its head sticking in the pore is simply larger than the number of ways it can find itself with the middle touching the pore.”
These theories of polymer networks have actually been around for a while. They were first proposed by the late Nobel laureate Pierre-Gilles de Gennes in the 1960s, and Bertrand Duplantier made key advances in the 1980s. Mihovilovic, Stein’s graduate student and the lead author of this study, says this is actually one of the first lab tests of those theories.
“They couldn’t be tested until now, when we can actually do single molecule measurements,” she said. “[De Gennes] postulated that one day it would be possible to test this. I think he would have been very excited to see it happen.”
Source: Brown University