Traditional drug design approaches largely rely on high-throughput screening, in which druglike compounds are tested against their target in large-scale assays to identify active hits. As these compounds are not specifically designed for the target in question, this leaves much to trial-and-error.
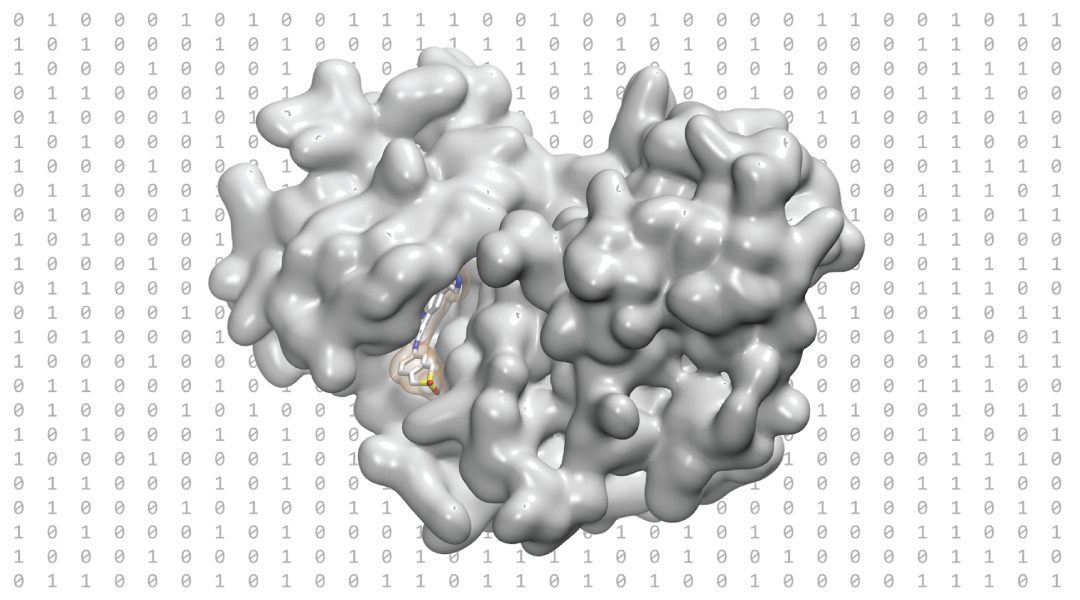
Deriving its name from the Greek “phoros” ( meaning a molecular framework that carries) and “pharmacon” (drug), a 3D pharmacophore is a three-dimensional, abstract representation of chemical and geometric features that characterize the molecular interactions necessary for a molecule to bind to a protein. Easy to use, pharmacophore models allow researchers to understand and interpret the chemical requirements for compound binding at a glance. Their intuitive and universal nature makes 3D pharmacophores the perfect tool for communication between researchers collaborating on interdisciplinary projects.
First conceptualized by Ehrlich at the end of the 19th century and solidified into its modern definition by Schueler in 1960, the notion of the 3D pharmacophore is not a new one. However, computational approaches allowing the application of 3D pharmacophores to virtual screening have revolutionized the field of rational drug design.
Physical compound libraries are limited by the number of compounds able to be physically synthesized and stored. In contrast, advances in machine learning have seen the generation of virtual databases containing billions of theoretically “synthesizable” molecules. Faced with this slew of potential hit compounds, 3D pharmacophores represent a unique opportunity to identify molecules fulfilling the specific molecular interactions necessary for binding to a certain target. In a process termed virtual screening, the 3D pharmacophore is used as a template to filter suitable compounds, introducing an unprecedented level of efficiency into the screening process.
In a recent study published in WIREs Computational Molecular Science, a team of researchers from Freie Universität Berlin explore the impact of 3D pharmacophores on drug discovery, as well as recent developments in the field.
Since traditional 3D pharmacophores only provide a simplified, static representation of molecule-target interactions, new directions in 3D pharmacophores are evolving towards including data from molecular dynamics simulations. These allow researchers to investigate the evolution of molecular interactions over time, providing an unseen level of detail on the binding event as a dynamic process. The deeper insight introduced by the inclusion of dynamics was used to rationalize differences in affinity between two agonists at the M2 receptor displaying the same static pharmacophore. Investigations into the binding dynamics of the two molecules revealed that the higher-affinity ligand adopts a different geometry within the binding pocket. This information can inform the design of high-affinity ligands for the receptor.
Pharmacophoric descriptors are increasingly being used as input in machine learning methods. Trained by pharmacophores describing known binding sites, the DeepSite machine learning model can detect unknown binding cavities by analyzing protein surfaces. KDEEP, a model able to predict the binding affinities of ligands in protein binding sites, is trained by protein-ligand complexes with known binding affinities depicted using pharmacophoric descriptors.
Pharmacophore approaches are no longer reserved for those with access to high-tech equipment. The advent of web applications enabling machine learning approaches, online pharmacophore generation, and virtual screening circumvent the need for high-performance computing clusters and license fees for academic researchers. This accessibility opens up the possibility of researchers generating and using pharmacophores from home, or in labs not specialized for computational approaches, at no extra cost.
As technology continues to evolve at a rapid pace, the implementation of machine learning, web applications, and dynamics promises to propel the already powerful tool of the 3D pharmacophore into the modern age, unlocking new perspectives on molecule-target interactions and paving the way for targeted and specific drug design.
Written by: Theresa Noonan
Reference: D. Schaller, et al. ‘Next generation 3D pharmacophore modeling‘ WIREs Computational Molecular Science (2020). DOI: 10.1002/wcms.1468