This ability to regulate sugar metabolism is just one example of the genetic switches discussed in a Review recently published in the journal WIREs Nanomedicine and Nanobiotechnology by Davey and Wilson at the Georgia Institute of Technology, titled “Deconstruction of complex protein signaling switches: a roadmap toward engineering higher-order gene regulators.”
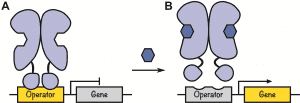
Cartoon schematic of the lac operon. A. Find out more here.
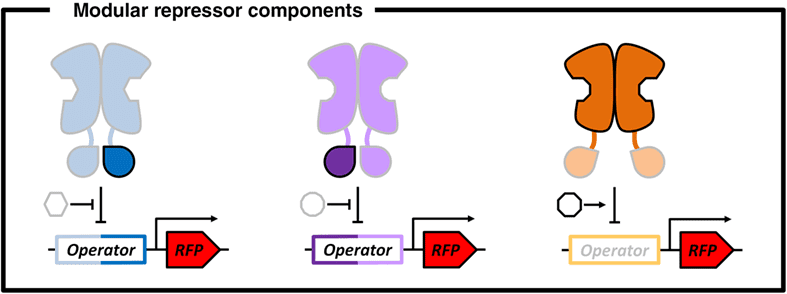
In their review, the authors survey a set of repressor proteins that have evolved in response to a variety of molecules and expand upon their utility as genetic switches. By scavenging components from these existing repressor proteins, new functions can be developed in the laboratory. For example, components belonging to LacI and GalR, two repressor proteins regulating the function of two operons in response to two distinct sugar molecules lactose and fructose (see Figure 2A), respectively, can be swapped to yield new repressor proteins capable of regulating sugar metabolism in response to each other’s respective sugar molecule (see Figure 2B).
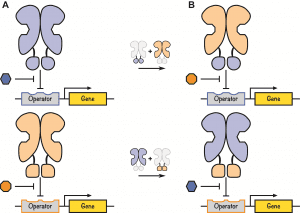
Cartoon representation of repressor design. A. Find out more here.
As a result, these repressor proteins can serve as the blueprints for new molecular machines and aid in the construction of synthetic biological circuits. The versatility of this regulatory machinery can be exploited to produce synthetic biological circuits that could in theory be expanded to the programming of specific cellular responses to multiple input stimuli. This idea can be conceptualized in a manner similar to that of a computer. By creatively recombining components of nature’s regulatory proteins, new orthogonal switches can be engineered. The nature of these novel switches will be a valuable tool for synthetic biology, bioindustrial, and biomedical applications in the near future.
Synopsis by James A. Davey and Corey J. Wilson, School of Chemical and Biomolecular Engineering, Georgia Institute of Technology