All cells in our body have been optimized over millions of years of evolution to deal with many environmental, disease, or external perturbations. Cells are intrinsically programed to activate a cascade of molecular responses that enact a complex but coordinated dance to express the proper functional molecules necessary to recover from the influences of specific environmental triggers. These cascades often directly regulate the synthesis of ribonucleic acid (RNA) from deoxynucleic acid (DNA) molecules present in every cell, with RNA molecules then acting as messengers that enable the expression of the appropriate protein pathways. Regulation of both the levels and composition of RNA molecules are crucial for establishing the molecular diversity necessary to adapt and survive a new environmental condition.
In a review in WIREs RNA, Pai and Luca discuss environmental influences on RNA processing, a step in the RNA maturation process comprised of a set of mechanisms that enable a cell to diversify the RNA molecules created from a single gene.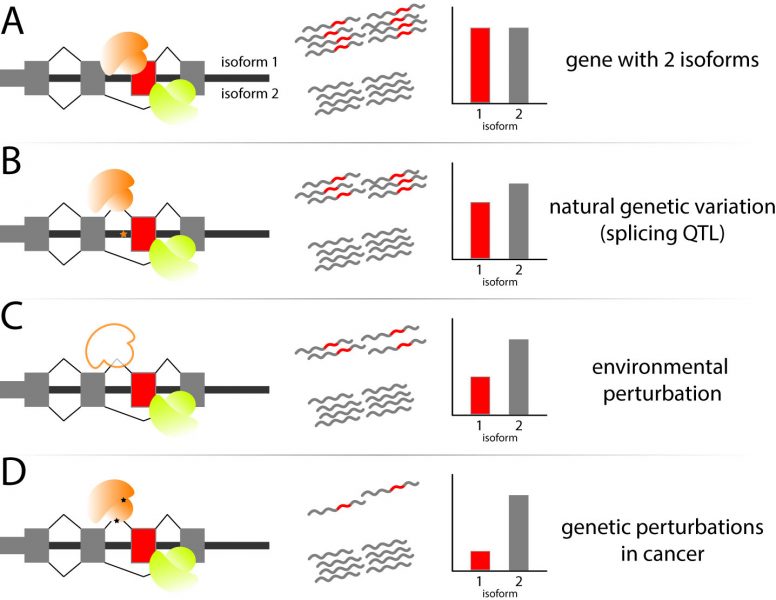
These mechanisms encompass the choices made in alternative transcription initiation, alternative mRNA splicing, and alternative cleavage and polyadenylation. The review focuses on our current understanding of how each of these mechanisms are dynamically regulated following well-studied environmental influences, including chemical treatments, the innate immune response, heat shock, DNA damage, and cancer progression. Growing scientific observations have made it clear that there is extensive remodeling of the composition of RNA diversity, with each perturbation enacting a specific set of coordinated changes unique to the functional or regulatory needs of the cell. In turn, these RNA processing changes regulate the levels of RNA molecules or the downstream levels or composition of proteins involved in cellular response.
Fine-tuned cellular responses are often dependent on the genetic makeup of cellular population—with known inter-individual variability underlying pharmacological efficiency, immune response phenotypes or cancer susceptibility. Pai and Luca discuss efforts to profile genetic loci that influence inter-individual variation in RNA processing and how these studies can be expanded to further understand the role of RNA processing in disease susceptibility and progression. Finally, the review discusses the technological advances that have enabled recent in-depth studies of RNA processing and transcriptome composition, while highlighting the challenges remaining in order to understand the causes and consequences of RNA processing dynamics upon perturbation of the cellular environment.