Chromosomal double strand breaks occur spontaneously during DNA replication. If left unrepaired, such breaks threaten genome integrity and therefore ultimately cell viability. To counteract these harmful effects, cells have developed intricate repair mechanisms. One of these repair mechanisms, namely homologous recombination, is the topic of a review by James Haber published in BioEssays.
The author discusses the mechanisms of homologous recombination and points out that the key step in this repair process is finding a homologous DNA segment that can be used as a template. The first step in homologous recombination is 5’ to 3’ resection of the double strand break to reveal single stranded DNA. The recombinase Rad51 subsequently forms a nucleoprotein filament on this single stranded DNA with each recombinase subunit binding three nucleotides. This Rad51 nucleoprotein filament is responsible for carrying out the search for homology.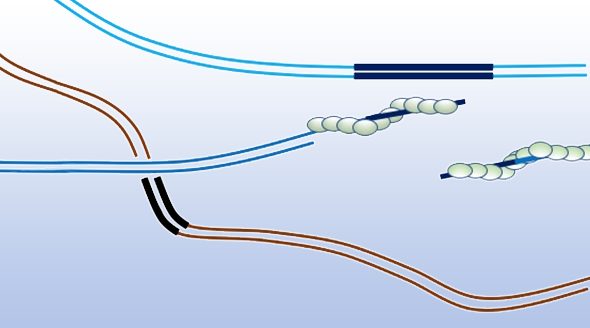
Finding a homologous sequence for repair depends on many different factors as pointed out by James Haber. Contact probability, that is the likelihood of these sequences finding one another, is one of these factors. This aspect is influenced by the physical proximity of potential homologous donor sequences in the nucleus, and in general, intrachromosomal repair is favored over interchromosomal repair. The length of the homologous sequence as well as the degree of homology are also important factors. Some in vitro studies have suggested that the minimum length of homology that will form a stable structure could be as little as six bases. And Rad51 seems to be able to tolerate a certain number of mismatches.
Further technical advances, e.g. in live-cell microscopy will hopefully help to reveal even greater detail in these early steps of homologous recombination. For more information, please also take a look at the video abstract where James Haber talks about his BioEssays review.
















