Image credit: Institute of Photonics and Electronics, Czech Academy of Sciences
Scientists have, for the first time, directly imaged fluctuations in the shape of single macromolecular species using a rather simple optical microscope.
“Ever since super-resolution microscopy revealed nanoscopic details about where in the cell specific single biomolecules do their job, we have dreamed about seeing the dynamics of the protein machinery which keeps life ticking,” says Marek Piliarik from the Institute of Photonics and Electronics in Prague.
Light microscopy offers insight into the dynamics of biological matter. However, the level of spatial details is intrinsically limited by the diffraction limit of light. Therefore the shape of sub-diffraction species can not be optically resolved.
In a study recently published in Small Methods, researchers introduce a new microscopy toolbox to discern the shape of protein structures.They employed the new method to visualize a series of individual conformational changes confined deep below the diffraction limit of light that characterizes the dynamics of single disassembling microtubules essential for cell function.
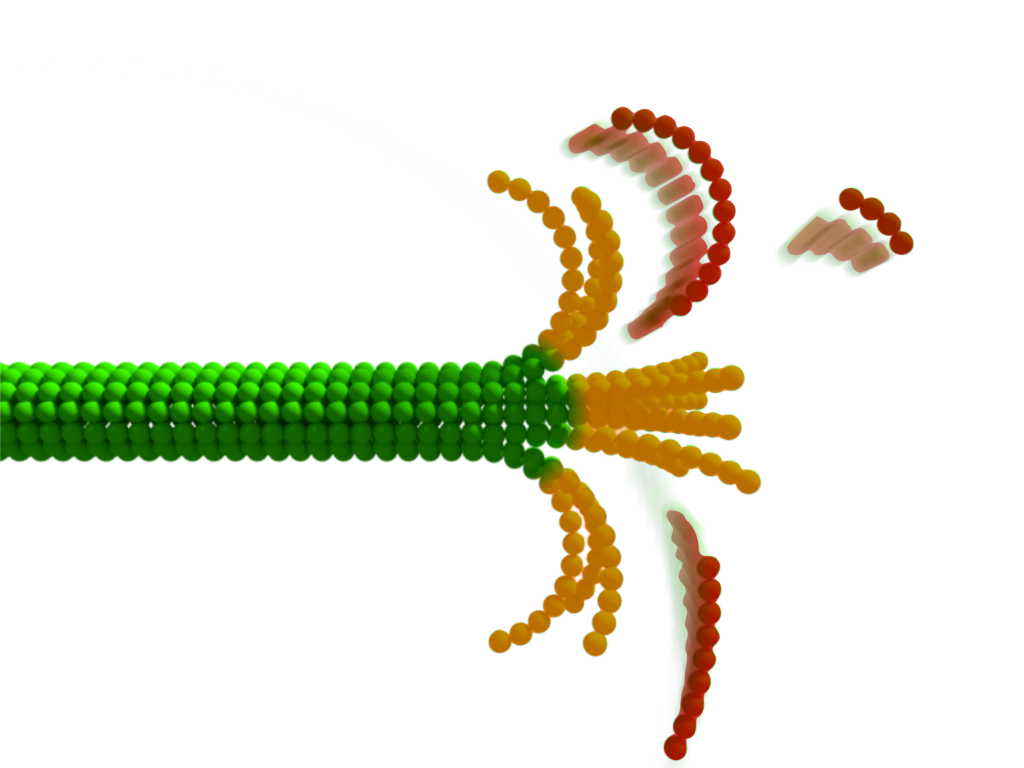
Microtubules are among the most dynamical cellular structures that perpetually grow and shrink,enabling vital processes in cells including division or motility. The presence of nanometer-sized curved structures at the tip of a disassembling microtubule observed on static electron micrographs have puzzled scientists for decades. To date, no time-resolved data has allowed them to gain an insight into the disassembly process.
Now, the group of authors from Prague have discerned the changes in the light scattering of protein structures owing to fluctuations in their immediate shape even though these transient segments are normally hidden in the diffraction-blurred edge of the microtubule. They were able to capture the choreography, rate, amplitude, and spatiotemporal displacement of transientstructural changes at the tip of disassembling microtubules.
Zdenek Lansky from the Institute of Biotechnology CAS adds a biological perspective to the work: “The unprecedented resolution of the method enables us to better understand the dynamic changes in the structure of microtubules — elusive mechanisms that underpin essential biological processes, such as cell division.”
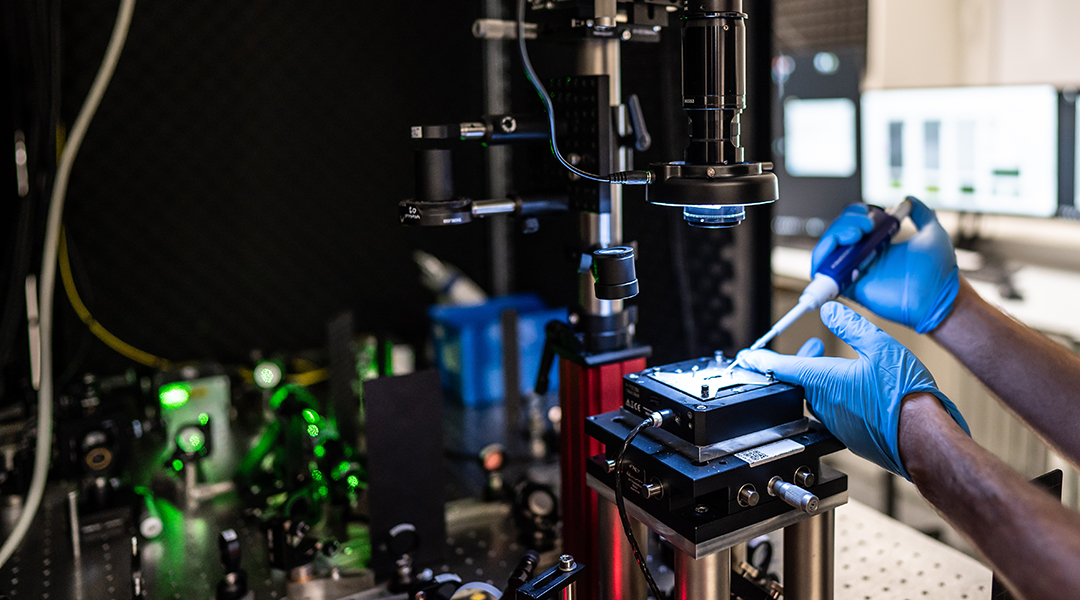
To achieve this, researchers adopted one of the most sensitive light microscopy techniques, recently coined sinterferometric scattering microscopy, which is ultimately capable of imaging single unlabeled proteins. The team from two institutes a tthe Czech Academy of Sciences pushed the limits of the technology beyond just detecting the light extinction signal cast by a structure of one or a few protein molecules in a split of a millisecond. The variation in a specific fraction of its signal allowed the scientists to resolve changes in the immediate shape of the protein structures.
“The fact that we can see protein-sized structures changing shapes only milliseconds before they break apart still amazes us. When considering the method’s capability of non-invasive imaging with up to MHz repetition rates, we believe it has significan tpotential to become a new window into cell machinery. The concept of seeing geometrical details of objects hidden in the diffraction bluris a greatopportunity for the whole field of optical microscopy,” said Milan Vala, the first author of the publication.
Piliarik shares his vision: “We believe that our technique will open a whole new field of applications similar to super-resolution microscopy that do not rely on a fluorescent probe to light up but simply resolve a particular conformation of natural protein structures.”
Reference: Milan Vala, et al., Nanoscopic Structural Fluctuations of Disassembling Microtubules Revealed by Label-Free Super-Resolution Microscopy, Small Methods, DOI: 10.1002/smtd.202000985
Press release provided by the Institute of Photonics and Electronics of the Czech Academy of Science