Morphogenesis is the series of organized and integrated processes that transform cells into functional organ systems during embryonic development or in adapting to environmental conditions. microRNAs are 21–25-nucleotide-long non-coding RNA molecules which function in transcriptional and post-transcriptional regulation of gene expression, and were recently implicated in the regulation of morphogenesis. Identifying and characterizing these microRNAs is essential for delineating the molecular events that drive this vital developmental process. Hence there is an urgent need for methods to assess microRNA function using various in vitro and in vivo experimental models.
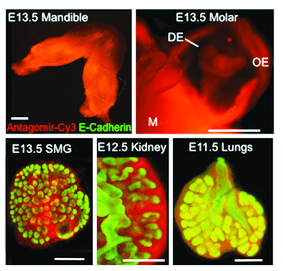
Epifluorescence images of various organ explants transfected using peptide based nanoparticles.
In the March Supplement of Current Protocols in Cell Biology, NIH investigator, Ivan Rebustini, describes a detailed protocol to assess microRNA function in explanted organs. This method functionally characterizes individual microRNAs during organ morphogenesis using peptide-based nanoparticle transfection to deliver oligonucleotides that act as microRNA inhibitors (antagomirs) or activators (mimics) inside embryonic explanted organs. Essentially, this system is designed on the impact of microRNA loss or gain of function on organ morphogenesis. The protocol also describes further validation of the microRNAs identified by the screen using a microRNA pulldown (miR-PD) assay to identify target genes associated with interesting and significant physiological functions.
This protocol has several advantages over other available methods: it is inexpensive, relatively straightforward and uses minimal culture medium, tissue sample and transfection cargo molecules (antagomirs and mimics). Multiwell analysis makes it high-throughput, which improves rigor and reproducibility.
The protocol covers assay limitations and provides a thorough Troubleshooting section with step-wise directions on how to address discrepancies and avoid artifacts. In addressing potential pitfalls, the author proposes some useful approaches to eliminate false positives or negatives. The protocol includes important recommendations, such as the use of tested and certified negative controls (off-target microRNAs), positive controls (microRNAs known to affect organ system), and testing of transfection cargo molecule concentrations to determine the limits of assay detection.
Easy adaptability, target specificity, simplicity and the high throughput of this method makes it a valuable strategy to replace traditional microRNA targeting using liposome-based transfections. Findings from this screen are relevant towards the understanding of embryonic development, and of organ regeneration, a field with tremendous therapeutic potential.
Visit Current Protocols in Cell Biology to browse cutting-edge as well as established protocols.