Cell migration from one area to another typically in response to chemical stimulus (chemotaxis) is essential for various physiological functions such as wound repair, cell differentiation and embryonic development. It also forms the basis of cancer metastasis wherein cancer cells migrate from the original cancer site and form secondary tumors. Hence, identifying inhibitors of cell migration and chemotaxis can provide insights into mechanistic studies for cell physiological functions and lead to novel cancer therapeutics. However, existing assays for cell migration and chemotaxis such as the monolayer scratch assay, transwell assay, and EZ-TAXIScan assay are not adaptable for high-throughput screening.
In the March 2017 Supplement of Current Protocols in Cell Biology, researchers Xin-Hua Liao from Fujian Medical University and Alan R. Kimmel from NIH describe a detailed protocol for a unique high-throughput assay to identify novel small-molecule inhibitors of cell migration and chemotaxis using Dictyostelium, an excellent model organism that shares cell migratory pathways with mammalian cells.
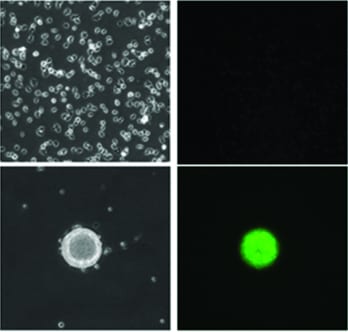
The a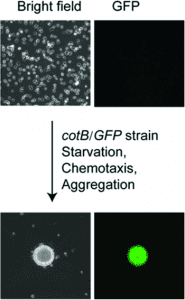 ssay principle is based on the cellular response of Dictyostelium to starvation which includes chemotaxis and cell migration followed by formation of multicellular aggregates. The authors describe the use of a GFP-reporter which is activated only in the multi-cellular agggregates resulting from cell migration. The GFP signal is measured using laser scanning cytometry. The protocol has addressed and resolved a major limitation of low-throughput current cell migration assays through automation and miniturization whereby an entire 1536-well plate can be assessed using only 8000 cells/well (∼6 μl) in just 10 minutes.
ssay principle is based on the cellular response of Dictyostelium to starvation which includes chemotaxis and cell migration followed by formation of multicellular aggregates. The authors describe the use of a GFP-reporter which is activated only in the multi-cellular agggregates resulting from cell migration. The GFP signal is measured using laser scanning cytometry. The protocol has addressed and resolved a major limitation of low-throughput current cell migration assays through automation and miniturization whereby an entire 1536-well plate can be assessed using only 8000 cells/well (∼6 μl) in just 10 minutes.
To avoid false negatives from single concentration analysis, authors describe a serial dilution method and to prevent false positives by agents that inhibit cell migration indirectly through cell killing they adapted a ATP content assay for measuring Dictyostelium viability.
In addition to providing thorough details of each step for these protocols, authors describe optimization of critical parameters such as cell density, incubation times and image detection consideration. Using descriptive figures and screenshots, the protocol also guides users through anticipated results and data analysis. This method was able to screen >10,000 compounds making it a valuable strategy for identifying novel anticancer therapeutics or studying factors and mechanisms that influence cell migration.
For new and established protocols in cell biology, visit Current Protocols in Cell Biology.