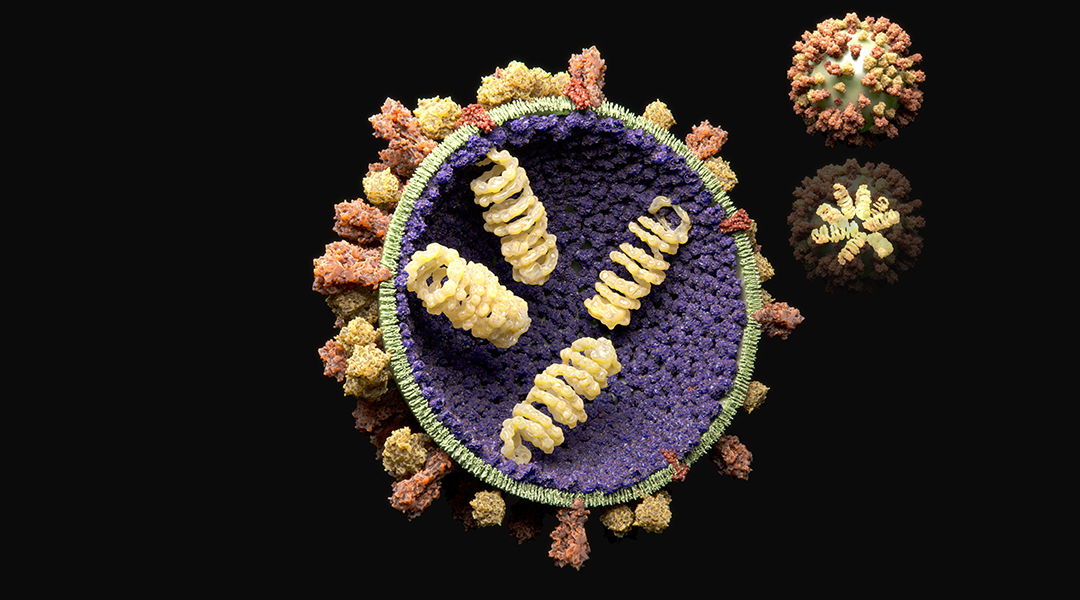
Image credit: Juan Gaertner/Science Photo Library
Understanding the intricate mechanisms behind viral infections is crucial to fighting them — as evidenced by the current global pandemic caused by the SARS-CoV-2 virus.
Intensive research efforts have gone into understanding how viruses infect and replicate within our cells. In RNA viruses, such as the poliovirus, rhinovirus (the cause of common cold), and aphtovirus (the cause of foot and mouth disease), infection occurs through a complex process that involves the creation of virions. These are particles that contain an outer protein shell called a capsid and an inner core that contains the virus’ genetic information and which these viruses use to continue infection and the spread of their genetic information.
While much has been elucidated about this process, according to Bogdan Dragnea, professor of chemistry and adjunct professor of molecular and cellular biochemistry at Indiana University, a longstanding question that researchers have grappled with is when virions form, how do they avoid encapsulating unwanted genetic material from the cell?
“It is still unclear how viruses differentiate between their own RNA and non-cognate (non viral) RNA,” said Dragnea. “It’s most likely a system-dependent answer, but when it comes to viruses, generalizing is a dangerous thing.”
In a study published in the journal Small, Dragnea and his collaborators Joseph Che‐Yen Wang and Martin F. Jarrold sought to understand how RNA viruses package their genetic information in order to further our understanding and help develop effective antiviral drugs, treatment strategies, and, of course, vaccines.
According to the researchers, the viral genome plays an active role in assembling these virions by recruiting “coat proteins” also known as virion proteins. These form the protective shell around genetic material inside the virus through spontaneous self-assembly. What is remarkable is that inside the crowded environment of a host cell, which contains all its machinery and countless biomolecules, viruses are effective at producing virus-like particles that avoid encapsulating smaller, non-viral molecules.
There are several hypotheses for how this happens. “For one, RNA and coat proteins have complementary structures, like a lock and key, that mutually encourage binding of the right RNA to to virus proteins,” said Dragnea. “This binding complex then attracts more proteins and guides them into a properly growing arrangement that ultimately results in a virus particle.
“Another possibility is that virus assembly occurs in the confines of virus-induced cellular compartments known as virus factories, which increase the effective concentration of those components that are required and restrict the access of those that are not.
“The third strategy comes from the interplay between the size of the RNA and that of the capsid. A preferred radius of curvature is encoded in the coat protein blueprint. If the RNA is too large or too small, the virus shell encapsulating it will be under elastic stress. These are three distinct strategies, but I would be surprised if they were mutually exclusive.”
Experimental as well as theoretical challenges have impeded our complete understanding of this process as its difficult to track. Until a few years ago, researchers would use averaging models and bulk kinetic studies to predict the structure of viral assemblies. “[These] relied on the assumption that a virus shell assembles through a series of protein addition steps to a growing intermediate fragment of the final structure,” said Dragnea.
To gain better insight, the team analyzed the assembly of brome mosaic virus (BMV) coat proteins and nucleic acid oligomers using a tandem approach that incorporated structural elucidation as well as mass analysis, which provided them with a previously unattainable clarity.
“It has been known for a while that the coat proteins of certain icosahedral RNA viruses can readily assemble around oligonucleotides and a broad variety of polyanions in vitro,” said Dragnea.
“However, the structures of virus-like particles formed in this way were not known. Our data suggests that in the presence of a heterogeneous nucleic acid population assembly [one that incorporates non-viral genetic material, for example], this may result in non-native cage structures, which are strained.”
To this end, using their tandem analytical technique the team was able to identify the formation of intermediate virion structures that differ from the well-known icosahedral shape commonly observed in these types of viruses.
“[These intermediates] may then disassemble easily into fragments that could be later used for growth into a native structure, should the right RNA displace the non-cognate one. This kind of pathway was excluded from models, and with a good reason — there was no data,” added Dragnea.
This means that next steps would involve creating models that incorporate more flexible, equilibrating subunits that don’t rigidly adhere to the final, native structure of the virion cage.
“I think the big challenge is to develop tools that follow dynamic processes inside the cell,” said Dragnea. “Improved computational and in vitro experimental single-particle approaches are valuable and will continue to provide useful insights. However, it would be nice to have methods that bridge computational, in vitro, and in vivo results. This is where fundamental research is going to be crucial.”
Reference: Kevin Bond, et al., Virus Assembly Pathways: Straying Away but Not Too Far, Small (2020). DOI: smll.202004475