A Trifunctional Linker for Purified 3D Assembled Peptide Structure Arrays: Epitope and technology overview
Research today is largely data driven, relying on high-throughput technologies where easy handling, low cost and miniaturization all play an important role.
To study interactions between biological molecules including proteins, microarrays provide a platform that fits the bill. However, especially high-density protein arrays that can be used to study interactions that depend on protein conformation are expensive to manufacture.
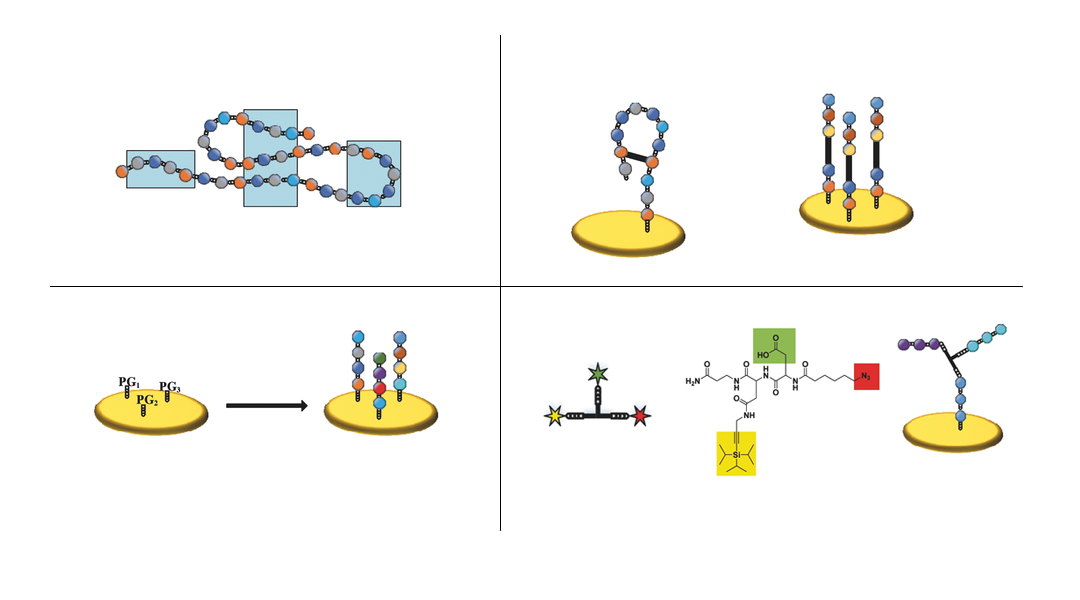
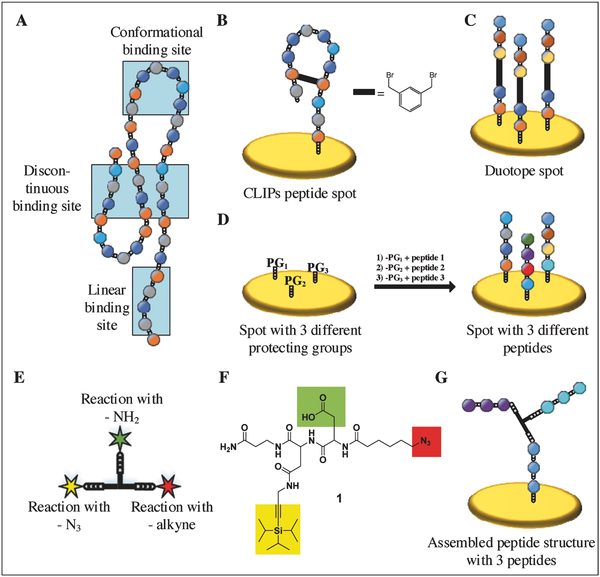
Researchers from the Karlsruhe Institute of Technology, Germany, developed a method to create arrays of branched peptide assemblies as a surrogate for protein arrays. While conventional peptide arrays permit the screening of linear and cyclic binding sites only, the new technology presented in Small Methods allows the researchers to study conformational and discontinuous binding sites on 3D assembled peptide structures.
Proteins can be “mimicked” by arranging several peptides at a defined distance to each other through the use of a trifunctional linker with an azide, a protected alkyne, and a carboxyl group. The linker can react with two or three different peptides and due to their spatial proximity, the peptides can assemble into peptide structures mimicking protein epitopes. The technology was demonstrated both on beads and on a polymer surface.