Scientists learned long ago that the flow of information from DNA to RNA to protein within a cell is a highly regulated process involving thousands of proteins in the cell working together to regulate the right genes at the right time. Once an RNA molecule is transcribed, it can be driven toward many different fates depending on the needs of the cell. The RNA can be quickly sent into the cytoplasm for translation, alternatively spliced to create different protein sequences, or sequestered for further processing only upon receiving cellular signals. These decisions about RNA fate are controlled through proteins that bind the RNA, known as RNA binding proteins (RBPs).
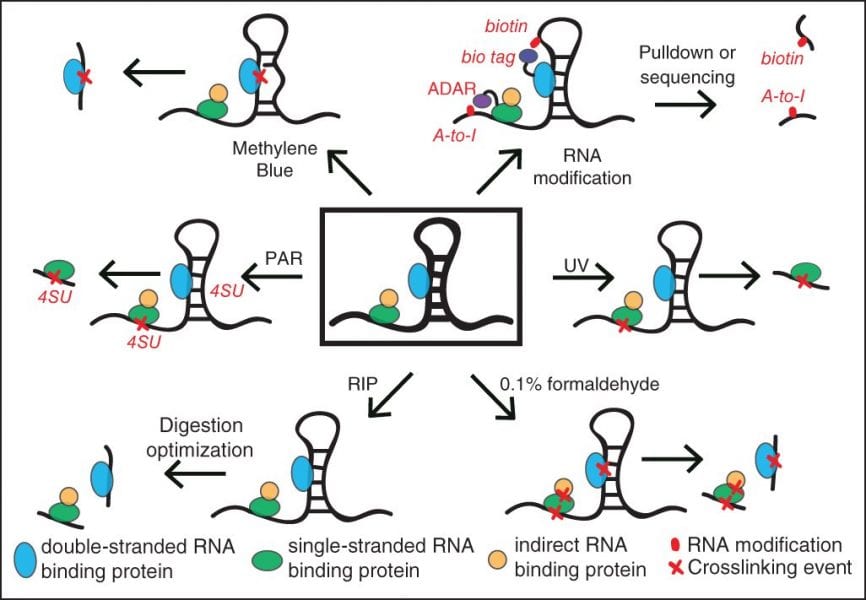
While it is known that RNA molecules are highly decorated by RBPs beginning immediately after transcription, much remains unknown about which proteins bind where, and how these binding events control RNA behavior. As many diseases are caused through mutations in RBPs or their RNA binding sites, methods to map all of the RNA binding sites for RBPs are critical to improve the understanding of how cells determine the fate of each RNA molecule that is produced. The RBP field was revolutionized by RNA-ImmunoPrecipitation (RIP) and CrossLinking followed by ImmunoPrecipitation (CLIP) technologies that enabled researchers to identify which RNAs are bound by a protein and what specific region of the RNA in a transcriptome-wide manner. However, these methods have been experimentally challenging to perform successfully, particularly at large scale and in tissues and other precious samples.
In a recent review in WIREs RNA, “Advances and challenges in the detection of transcriptome-wide protein-RNA interactions,” Wheeler et al. explain the technical limitations of RIP and CLIP approaches that have historically limited the scope of identifying protein-binding sites on RNA. Most notably, isolation of the bound RNA fragments and the conversion into cDNA for a sequencing library have suffered from experimental inefficiencies. The authors highlight improvements in CLIP methods that incorporate novel ligation strategies and optimized reaction conditions to dramatically improve the experimental success rate. They also discuss a variety of techniques to isolate RNAs that are bound by proteins, including removing the requirement for RBP-specific antibodies and increasing the capture efficiency of structured RNA targets. Thus, this review provides both a general overview of RBP target identification as well as a deep dive into alternative strategies for key challenging steps, providing an invaluable resource for researchers deciding on how best to study the targets of an RBP of interest.
Kindly contributed by the Authors.