 Natural macromolecules such as enzymes and proteins effectively fulfill their function because of a precise, dynamic three-dimensional architecture – the quarternary structure. The foundation of these structures is nature’s ability to exactly control the amino acid sequence when building up proteins or enzymes in the ribosomes.
Natural macromolecules such as enzymes and proteins effectively fulfill their function because of a precise, dynamic three-dimensional architecture – the quarternary structure. The foundation of these structures is nature’s ability to exactly control the amino acid sequence when building up proteins or enzymes in the ribosomes.
Taking nature as a paradigm is a promising strategy, certainly also in macromolecular design. Since true sequence control lies still far beyond the scope of synthetic chemistry, an alternative approach towards artificial proteins is the folding of single polymer chains at predefined positions. Thus, mimicking their three-dimensional structure can be achieved on a very simple level. Analogous to proteins, such single chain architectures have been realized using covalent, dynamic covalent and supramolecular binding motifs. Nonetheless, up to now one important type of linkage that is also common in proteins and enzymes has been neglected: metal ion complexation.
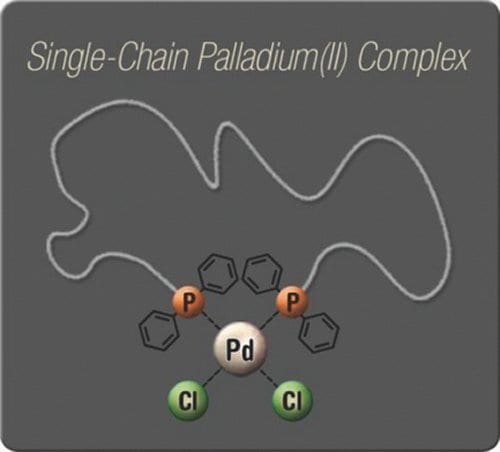
To address this void, a first example of metal-ligand interaction driven single chain self-folding is presented. The group of Christopher Barner-Kowollik (KIT, Germany) synthesized bistriphenylphosphine macroligands by a combination of controlled/living radical polymerization and modular ligation chemistry. In high dilution these bidentate macroligands form cyclic complexes with palladium(II) ions. The displayed fashion of folding single polymer chains can in the future be combined with other types of linkages, extended to other metal-ligand systems or even be a potential mimic of the active site of an enzyme.