Our DNA is a linear sequence of genes interspersed with long non-coding regions, comprising about 98% of our genome, whose function is only partially understood. In the last few years it has been discovered that non-coding DNA contains key regulatory regions that control gene activity through the formation of physical contacts with their target genes, thereby establishing cell-type-specific three-dimensional (3D) conformations of chromosomes.
New technologies, such as Hi-C, GAM or SPRITE, and developments in microscopy have revealed the sheer complexity of the genome 3D architecture and its deep connections with gene regulation. Importantly, it has been discovered that disruptions of chromosomal contacts induced by mutations, such as structural variations, are linked to diseases, including congenital disorders and cancer. However, the molecular mechanisms underlying chromatin folding remain largely unknown.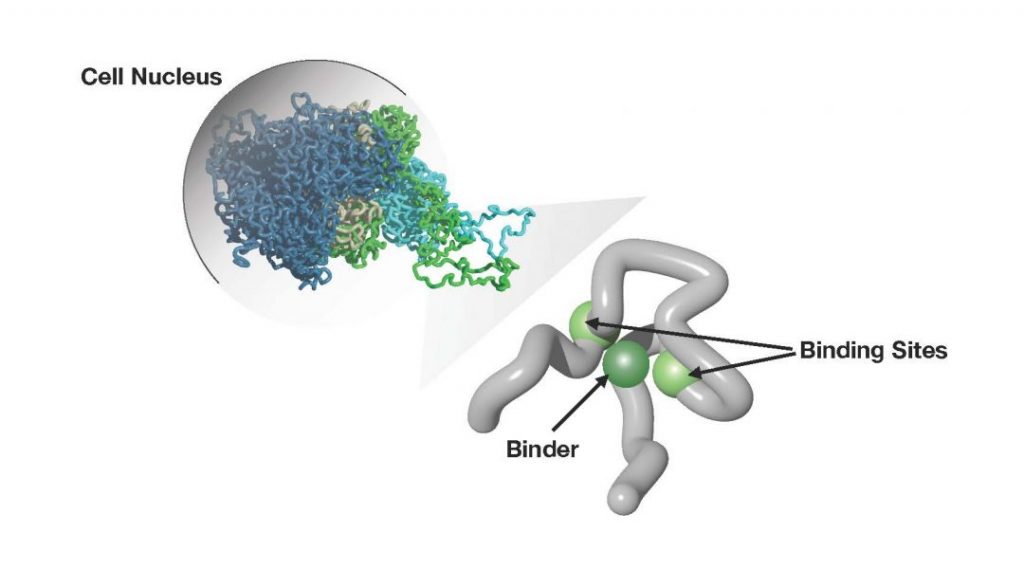
To make sense of the complexity of high-throughput contact data, principled approaches from polymer physics have been developed. They aim to dissect the molecular factors controlling chromatin folding, and transcription, and to explain the formation of chromatin structures at the single cell level, hence enabling the prediction of the impact of mutations. Some of the recent advancements in the field have been summarized, focusing, in particular, on the strings and binders and the loop extrusion models, and their comparison against experimental data.
The strings and binders considers the classical scenario of molecular biology where contacts between distal DNA sites and loops are established by diffusing molecules such as transcription factors, via mechanisms of equilibrium polymer thermodynamics. The loop extrusion model focuses on another classical scenario where loops are formed by molecules that bind to a site along DNA and extrude a loop, via off-equilibrium mechanisms.
Kindly contributed by the Authors.

















