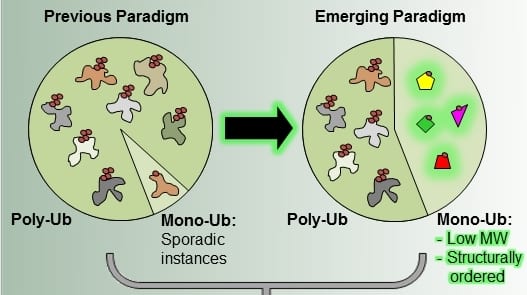
Polyubiquitination has long been known as the “canonical” signal targeting proteins for degradation by the proteasome. However, recent systematic screens in both human and yeast cells have revealed that the situation is far more complex than previously thought. Aaron Ciechanover and colleagues were able to show that a large number of proteins do not require a polyubiquitin chain to mark them for degradation. Instead, these proteins are degraded upon monoubiquitination. Interestingly, about 40% of mammalian and 20% of yeast proteins fall into this category.
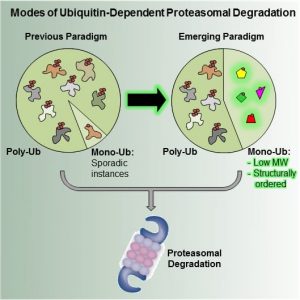 But what determines the type of signal used to mark a protein for degradation? As Livneh et al. point out in their review in BioEssays this is to a large extent determined by the characteristics of the protein itself. In general, for smaller proteins (up to about 150 amino acids) monoubiquitination is sufficient to target them for degradation whereas larger substrates require longer chains. Furthermore, at least in humans, monoubiquitinated proteins are less disordered than their polyubiquitinated counterparts. The authors suggest that polyubiquitination provides higher affinity and better binding to the proteasome and therefore it is not surprising that larger, more disordered proteins will need a longer ubiquitin chain to efficiently target them for degradation.
But what determines the type of signal used to mark a protein for degradation? As Livneh et al. point out in their review in BioEssays this is to a large extent determined by the characteristics of the protein itself. In general, for smaller proteins (up to about 150 amino acids) monoubiquitination is sufficient to target them for degradation whereas larger substrates require longer chains. Furthermore, at least in humans, monoubiquitinated proteins are less disordered than their polyubiquitinated counterparts. The authors suggest that polyubiquitination provides higher affinity and better binding to the proteasome and therefore it is not surprising that larger, more disordered proteins will need a longer ubiquitin chain to efficiently target them for degradation.
Overall, these findings have led to an expansion of the ubiquitin code, revealing that there is no single signal marking a protein for degradation. Protein degradation is rather achieved via an intricate interplay involving many factors: from the characteristics of the substrate itself to the different enzymes involved in this whole process.
Also watch the video abstract where Aaron Ciechanover talks about these recent findings.