Cells express their genes by transcribing a messenger RNA (mRNA) copy from DNA. These messages are then translated to make proteins, the functional workhorses of the cell. If cells determine that an mRNA has an error or is no longer needed, they use specialized pathways to degrade it. One such pathway, called nonsense-mediated decay (NMD), is tasked with recognizing mRNAs that code for proteins that are too short as a result of sequence errors. The challenge for the pathway is accuracy: how can it correctly identify the mRNAs that should be degraded from the bulk of mRNA in the cell?
As might be expected, a complicated challenge requires a nuanced solution. In their review in WIREs RNA on NMD, Kishor and colleagues highlight the features and structure of an mRNA that would lead to its degradation. These include whether or not the mRNA is being actively translated into protein, as well as what proteins are bound to the mRNA in question. Some mRNAs bind proteins that mark them for rapid degradation, while others recruit protein shields that protect them from recognition by NMD. The end result is a pathway that is highly evolved to target the right mRNAs for destruction while allowing the rest to continue making functional proteins.
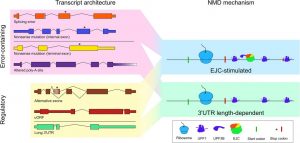
The ribonucleoprotein (RNP) configuration of the 3′UTR of a message influences substrate selection for NMD. Transcripts identified as decay substrates by the NMD machinery may contain errors or may be a part of a cellular regulatory program. The trigger for decay, however, is not determined by the process that generates a transcript but rather its 3′UTR RNP composition.
Human health depends on NMD getting this right. Documented pathology that results from NMD failure includes muscular dystrophy, intellectual disability, and cancer. Embryos without functioning NMD are not viable. As continuing scholarship furthers our understanding of the interplay between pro- and anti-NMD signals, we will better be able to predict targeting and harness NMD to promote health while preventing disease.
Kindly provided by the Authors.