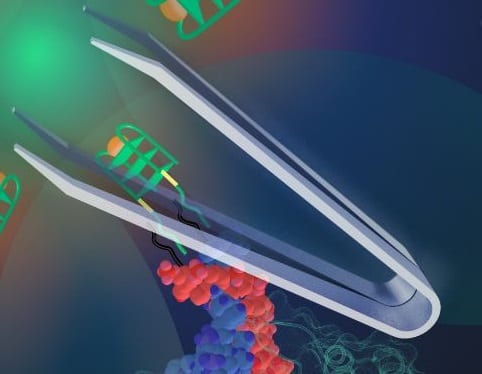
The elegant system of complementary base pairs that characterize DNA has the potential to lead to wide variety of assembly mechanisms and shapes that can be exploited to build complex nanostructures. DNA can be folded into quadruplex structures, which can be combined with porphyrin hemin to result in catalytic peroxidase activity that is amenable to a colorimetric response. By developing an approach that triggers DNA to adopt such a quadruplex structure, it is possible to design a dynamic response system, which is sensitive to the presence of a particular molecule and can be used in diagnostic or sensing applications. Such a dynamic response has been shown using so-called DNA tweezers, a DNA nanomachinery that closes in response to the presence of a single-stranded nucleic acid using complementary base pairing. Until recently, extending this method to sense whole proteins remained a challenge. However, in a study published in the new journal Advanced Biosystems, scientists from the University of Hong Kong demonstrated that it is possible to use a DNA aptamer split across a tweezer such that the presence of the target protein triggers the DNA tweezer mechanism.
As a proof of principle, the team led by Dr. Julian Tanner demonstrated the detection of malaria biomarker Plasmodium falciparum lactate dehydrogenase (PfLDH). Several critical factors had to be considered in order to realize this new nanotweezer machinery; the position of the split on the aptamer, the extent of self-complementarity, and the spatial flexibility for PfLDH binding all need to be precisely controlled. Optimization of these factors enhanced the binding affinity of the aptamer tweezers, ensured specificity to PfLDH, and resulted in a specific colorimetric response. This advance demonstrated the use of split aptamers for DNA nanostructures where protein sensing is a key component of an artificial biosystem and opens up new possibilities for the design of functional and responsive nanosystems.