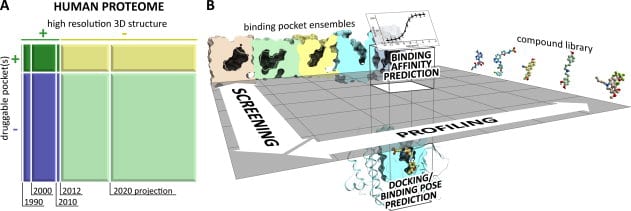
In this rapidly shrinking world with easy availability of air travel linking remote regions and the constant rise of drug resistant bacteria and viruses, an increasing number of new and previously tackled diseases are threatening human life and wellness all over the world. In response, pharmaceutical drug research for vaccines and therapy has had to come up with entirely new paradigms for rapid study of candidate drug molecules. The computational study of protein ligand interactions is crucial as it reduces the number of molecules that need to be tested in vivo and ultimately undergo drug trials. In recent years, there has been a rapid growth in the availability of crystallographic information about proteins and binding pockets. About 20% of the human proteome may have druggable pockets, for which structural information is presently available. A more accurate computational strategy involving quantum chemical methods is expected to provide more precise solutions.
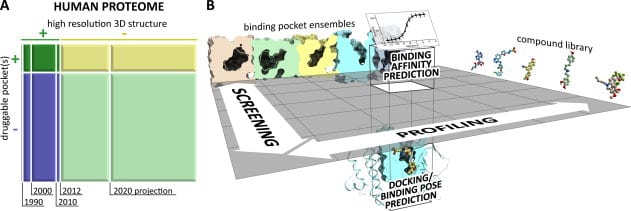
Typical steps in computational analysis involved include: (i) compound docking or pose prediction, (ii) compound screening, (iii) compound profiling, and (iv) compound binding affinity prediction. Most computational chemistry applications are centered on two critical steps performed for each compound and pocket structure – conformational sampling and scoring of obtained poses. A vast majority of existing work on conformational sampling involves the use of molecular dynamics and Monte Carlo methods and is quite mature. Scoring of poses can be done in a multitude of ways, including force-field methods that attempt to mimic the accuracy of ab initio quantum mechanical calculations and their predictive value. While inaccuracies exist in application of force-field methods, the insufficiency of the predictive power of structure-based computational approaches arises from incompleteness of input data. Experimental access to all binding site conformations is impossible. Compositional and conformational ensemble analysis is a practical approach to cover such gaps in data.
In a perspective article now available for free in the International Journal of Quantum Chemistry, Dr. Irina Kufareva and her colleagues from UC San Diego discuss quantum mechanical contributions to drug research in some detail. Covalent and non-covalent interactions can both be studied at a very high level of accuracy, but owing to complexity of calculations, systems with no more than about 70 atoms can be studied. Further complications arise from the need to add in entropic information through analysis of conformational ensembles. A hybrid approach that combines quantum mechanical calculations and molecular dynamics is more promising and was able to score more than 75% of the binders in one such study. Another alternative, semiempirical quantum mechanical calculations, attempt to reduce computational cost by reducing the basis set and simplifying the integrals. Recent attempts to model parts of interacting groups with simple molecules represent a promising first step in its application to this problem.
Approaches based on fragmentation of the system and decomposition of interactions have produced reasonable agreement with experimental results. Use of quantum mechanics based partial atomic charges has been seen to improve agreement with experimental values, provided solvation is accounted for. Coupled with availability of faster hardware, use of computationally expensive quantum mechanical approaches has become more popular in this field, as evidenced by the rise in number of publications devoted to the protein-ligand problem. Subject to the availability of accurate input data and errors in calculation, this trend is expected to continue and provide an additional supplementary approach to computational drug research.