In a new review published in WIREs RNA, the authors summarize recent progress of deep learning methods in predicting RNA–protein interactions, which play an essential role in many biological processes, such as post-transcriptional regulation of genes.
Dysregulation of RNA-binding proteins (RBPs) may lead to various diseases. Compared to experimental detection of interactions between RNA and proteins, computational methods are more efficient and time-saving.
With increasing high-throughput sequencing, more and more data related to RNA–protein interactions is accumulated, including RNA–protein complexes and cross‐linking and immunoprecipitation followed by sequencing (CLIP-Seq) data.
This data provides a large number of verified RNA-protein interactions as training information for machine learning models—especially data-driven deep learning models.
Deep learning has achieved remarkable success in many applications, including computer vision, speech recognition, and language translation. Thus, deep learning has also been applied for predicting RNA–protein interactions.
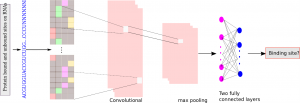 The authors introduce the databases of RNA–protein interactions, which can serve as building a training data set for deep learning.
The authors introduce the databases of RNA–protein interactions, which can serve as building a training data set for deep learning.
The authors also point out that RNA–protein interaction predictions can be formulated into three types of classification, including binary classification, and multi-label classification.
Additionally, the authors provide an overview of the successful implementation of various deep learning approaches for predicting RNA–protein interactions, mainly focusing on the prediction of RNA–protein interaction pairs and RBP binding sites on RNA.
According to the reported performance in the same data set, the authors found that deep learning is better at predicting RBP binding sites on RNA and yields a much better performance than traditional machine learning-based methods, due to the large volume of available training data.
To date, deep learning is still used for feature extraction for predicting RNA–protein interaction pairs, and cannot be trained in an end-to-end way due to rarely available RNA–protein pairs.
The authors additionally discuss the challenges and potential for improving the prediction methods for predicting RBP binding sites.
Kindly contributed by the Authors.

















