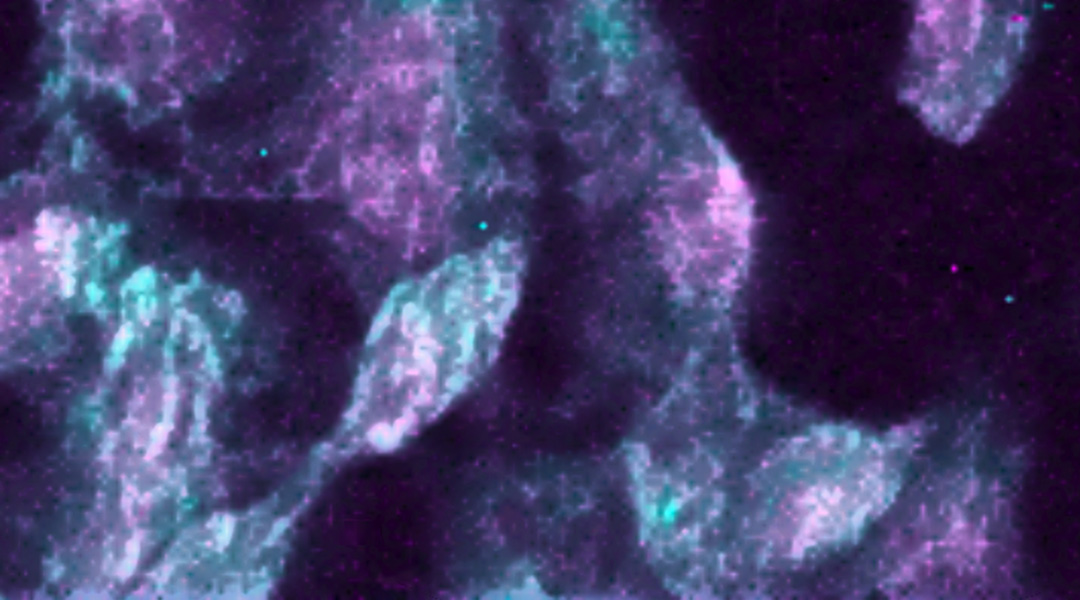
A microscopic molecular Raman image of cells infected by the SARS-CoV-2 virus. Image credit: Giuseppe Pezzotti
The continuing emergence of new viral variants of SARS-CoV-2 remains a strong likelihood while global vaccine inequity remains an issue. With mutations in the virus’ genome comes the possibility that vaccines will be rendered ineffective as the virus “learns” to evade immune responses.
To stay on top of the situation, scientists are constantly monitoring for new mutations through genetic sequencing using samples collected from infected patients. While this method is accurate, it can be time consuming and expensive.
An alternative, time-saving approach has been proposed by researchers at the Kyoto Institute of Technology and Kyoto Prefectural University of Medicine based on Raman spectroscopy and the ability to ID variants based on bar codes derived from their spectra.
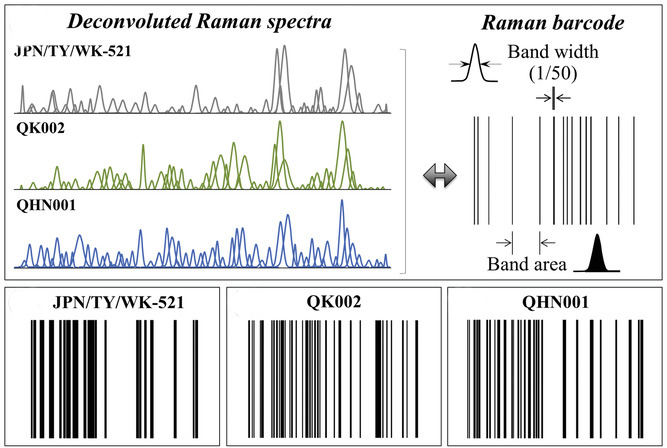
“Raman spectroscopy is a useful method in microbiology, capable of bringing insightful information at the molecular level,” explained Giuseppe Pezzotti, co-lead author of a recent study published in Advanced Science. “It can be used to follow in situ the metabolism and physiological changes of living cells, including the molecular details of their viral infection [see article’s feature picture]. We also have data on different influenza strains, and their inactivation mechanisms collected using Raman spectroscopy.”
Pezzotti and his colleague Osam Mazda say they realized an immediate application for their methodology with the onset of the current pandemic and hoped to provide useful information to help scientists get the situation under control.
Identifying the virus’ changing structural elements
Raman spectroscopy is a non-destructive analytical technique that provides information about chemical structure. A sample is irradiated with monochromatic light, usually from a laser, and the scattered light is collected and analyzed. A majority of the scattered light is the same wavelength as the incoming laser light, but a small portion is scattered at a different wavelength as a result of it having been absorbed and part of it re-emitted by chemical bonds in the molecule.
This is called “Raman light” and it is affected by the weights of atoms, the strength of their chemical bonds, and their spatial arrangement in a molecule. The easiest comparison for this is a “ball and spring analogy” in which bonds between atoms can be thought of as a spring connecting two balls. The frequency of vibration, which depends on the stiffness of the spring, how heavy the balls are, and how they are connected to the spring will change how they “bounce” and re-emit kinetic energy put into the system.
In a similar way, when light or energy is provided at the correct frequency, a chemical bond will absorb that energy and re-emit part of it as light. Depending on the previously mentioned factors, different molecular elements will scatter the incoming light in a specific way, emitting a unique Raman spectrum. As a result, Raman spectra provide a chemical fingerprint, which Pezzotti and Mazda have directly linked to the molecular structure of different SARS-CoV-2 variants.
“At the molecular level, there are a number of structural details that differ in [SARS-CoV-2] viral variants,” said Pezzotti. “These include amino acid residues, RNA bases, and secondary structure of proteins, and they appear clearly in the Raman spectrum.
For example, sulfur is found in the amino acids, methionine and cysteine — both found in viral proteins — and forms strong bonds with carbon atoms, making them clearly visible in Raman spectra at different frequencies depending on molecular structure and symmetry. The team was able to use these chemical signatures, which appear differently depending on the virus’ molecular structure, to note key differences between variants they were studying. “The differences in methionine and cysteine molecular symmetry of different SARS-CoV-2 variants were stunningly bold,” added Pezzotti.
Additionally, the team found they could use the fingerprints of RNA bases purine and pyrimidine to help differentiate between viral strains. “These bases possess molecular vibrations that are different to each other due to differences in their aromatic ring structures,” explained Pezzotti. “Characteristic vibrations, and thus Raman signals, can be located for each RNA base in the genome, with their relative intensities partly reflecting their statistical frequency in the RNA, but also sensitive to their sequence order.”
Creating barcodes for different variants for efficient record keeping
Combining clues provided by different structural elements, the team was able to infer changes in the final spike protein structures and identify elements in the viral genome that had “reshuffled” between variants. So far, the team has collected Raman data on a number of known SARS-CoV-2 variants, including the Delta variant.
The next step was assigning “Raman barcodes” to each variant depending on their spectra. This was done using an algorithm to assign each band in the spectrum a line of thickness equal to 1/50 of its bandwidth and a distance from the successive line proportional to the band area. This is just one of “many ways to build a barcode or even a QR code from a Raman spectrum,” said Pezzotti.
The goal is to establish an efficient, electronic means of record keeping and an improved ability to trace the many structural characteristics of variants encrypted in Raman spectrum.
“The capacity of Raman spectroscopy to both resolve the viral structure on a molecular scale and quickly provide information about new coronavirus variants could be key in future virology research and viral tracking,” added Mazda. “Beyond conventional PCR tests, Raman spectroscopy could enable fast on-site variant identification, provided that it could be coupled to technologies enabling quick and efficient virus isolation and concentration from clinical samples.”
Reference: Giuseppe Pezzotti, et al., Raman Molecular Fingerprints of SARS-CoV-2 British Variant and the Concept of Raman Barcode, Advanced Science (2021). DOI: 10.1002/advs.202103287