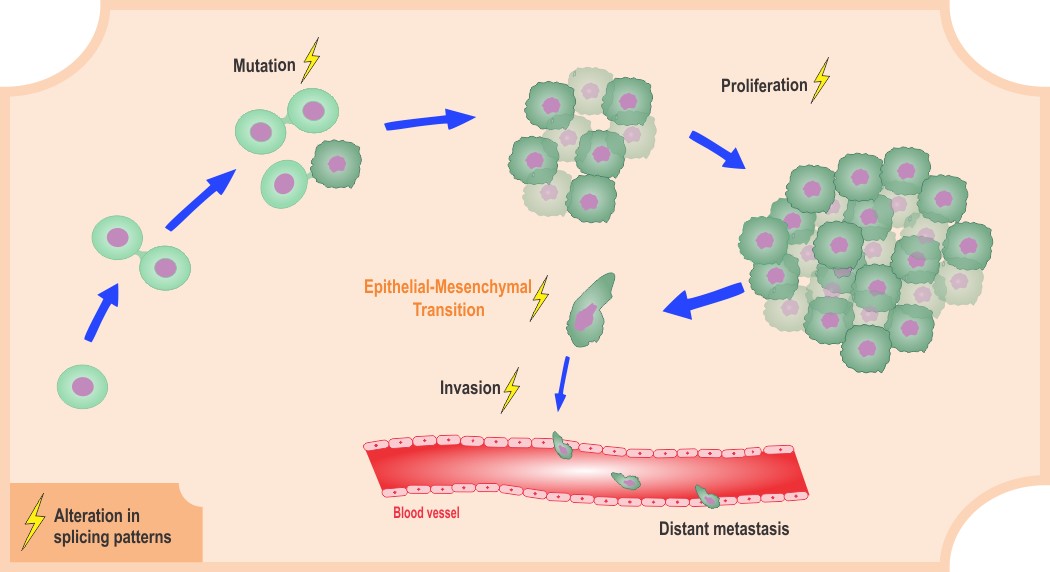
Cancer is a complex multi-factorial disease that results from many molecular alterations. Among the observed effects are alterations in proliferation, growth, replication, and death of cells. Global genomic analyses coupled to functional assays have shown that altered gene expression is one possible mechanism of disease development. In human cells, genes are decoded in immature messages, which must be processed to generate functional proteins. Gene expression pathways involve several control steps; DNAs transcribe RNAs, which are composed of sequences that code proteins (exons) and intervening sequences (introns). Splicing is an important step during RNA maturation that promotes intron removal and exon ligation, ultimately defining which information will be expressed by one particular cell type. Regulation of this processing step is critical, for instance, in the maintenance of cell homeostasis. Splicing can drive important gene expression changes, which may or may not lead to disease development.
Many molecular changes observed in cancer are a direct consequence of altered gene expression caused by changes in splicing regulation. Mutations in pre-mRNA splice sites, mutations in spliceosome components or regulatory proteins, as well as altered levels of specific proteins, can lead to alterations in the splicing process. Importantly, alterations in splice site preferences can generate alternative isoforms, enabling different biological functions, and might also be responsible for non-functional isoforms that can eventually cause dysregulation in cellular processes.
A WIREs RNA review by Patricia Coltri, Maria Gabriela dos Santos, and Guilherme Henrique da Silva discusses the major splicing changes observed in cancer cells, from mutations in pre-mRNA sequences to mutations in spliceosome components and alterations of protein levels that might promote splicing dysregulation, and ultimately lead to cancer. Based on the molecular changes, possible therapeutic approaches are also discussed. Splicing modulation is currently a promising and exciting therapeutic strategy, and it has opened a new field of research. The study of the mechanism of action of some drugs derived from bacteria have revealed spliceosome components as potential targets. Future biomedical approaches might confirm whether the use of these drugs can control splicing efficiency and the rise of related diseases.
Kindly contributed by the Authors.