The central dogma of biology describes the flow of genetic information from DNA to RNA to proteins. While RNA was originally believed to be a carrier of genetic information, subsequent work has shown something completely different: RNA is now known to have function independent of proteins, with a rich layer of regulatory networks. In fact, a large amount of the RNA present in a cell does not actually make proteins. This increased appreciation and understanding has led to many fascinating mechanistic insights into RNA and its role as a central player in cellular regulation and human disease.
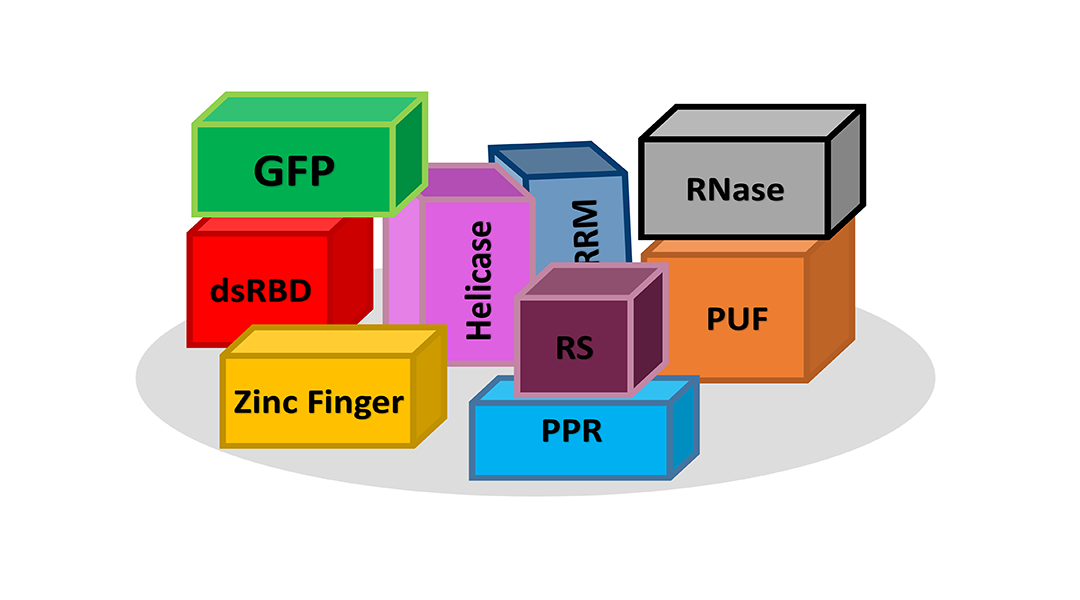
Helping to facilitate RNA function are a large number of proteins that can bind to and regulate RNA. These RNA-binding proteins, or RBPs, number in the thousands and are made up of many different independent modular segments similar to a child’s set of building blocks. In much a similar fashion, these “blocks” or domains provide nature with a way of mixing and matching different domains to generate new functions. In recent years, researchers have sought to learn from biology and use these building blocks to engineer new proteins with unique functions that are helpful in research and human health. In a recent study published in WIREs RNA, Professor Andrew Berglund and his colleagues describe recent advances and challenges in engineering RBPs.
“Engineering [RBPs] is a powerful tool for researchers to probe the mechanisms of RNA processing pathways” says Dr. Andrew Berglund, the newly appointed Director of the RNA Institute at the University at Albany. “It is also a promising approach for the development of novel therapeutic molecules.”
Potential targets for this approach are abundant as many human diseases have a strong RNA or RBP component, including the most common cause of muscular dystrophy and amylotrophic lateral sclerosis. For therapeutic purposes, engineered RBPs can be designed to replace a defective cellular RBP or bind and destroy toxic RNA. RBPs can also be engineered with new functions and/or targets as well as being marked or “tagged” so that the researcher can follow their progression within the cell, like a GPS tracker for RNA. The modular nature of RBPs makes it possible to add or mix function to suit the goal of almost any researcher.
In their study published in WIREs RNA, the research team highlights two specific types of RBDs domains, PPR and PUF domains, which are the most straightforward choice for protein engineering. Researchers have studied these domains, understand how to design them to bind specific RNA sequences, and even have websites that can be used to design a domain to target your RNA of choice. Not all RBP engineering is this straightforward, with most researchers having to consider other factors such as the type of linker between domains, where in the tissue or the cell that protein must go, and how to attach other domains to give the engineered protein function.
Ultimately, as more functions of RNA are discovered and more diseases are linked to RNA misregulation, the greater the importance will be for designing, engineering, and testing novel RNA binding proteins. Somewhere in a jumble of RBP building blocks may lie the key to unlocking the next big discovery on RNA and potentially the next generation of therapeutics to improve human health.
Kindly contributed by the authors.