Do not expect RNA to sit still. Although structural investigations typically illustrate a single, more or less static picture of an RNA, most RNAs most undergo conformational changes to exert their cellular function.
In a recent Open Access review in WIREs RNA, Dagenais et al. from the Université de Montréal, Canada report on how they gained novel insights into RNA dynamics by comparing structures of different conformational states. Their review is based on several recent high-resolution structures of the Neurospora Varkud satellite ribozyme. NMR structures of several individual subdomains of this catalytic RNA were determined as part of a divide-and-conquer approach to generate a three-dimensional model in solution. In addition, crystal structures were determined for active-site variants of the complete ribozyme that were trapped in a pre-catalytic state. By comparing the NMR and crystal structures, the authors present a detailed and complete portrait of the structural and dynamic properties of this catalytic RNA. In general agreement with previous biochemical and biophysical characterizations of the VS ribozyme, these recent structural studies provide a coherent atomic view into how the ribozyme recognizes and cleaves its stem-loop substrate.
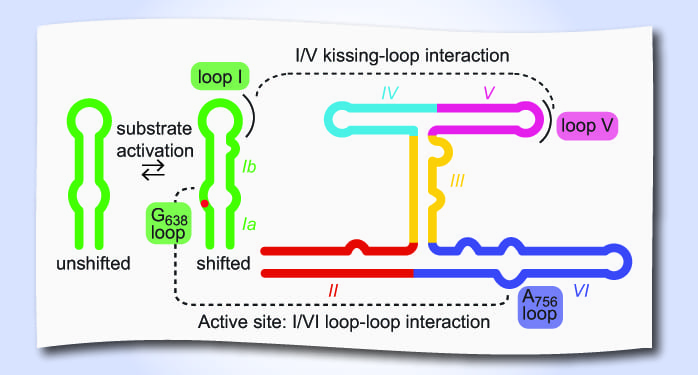
Like many RNAs, the VS ribozyme dances to its own tune, undergoing dynamic exchange between multiple conformational states before cleaving its substrate. A few well-choreographed moves bring the ribozyme to the ultimate cleavage step: recognition of its substrate by a kissing-loop interaction followed by activation of the substrate via helix shifting and formation of the active site via a loop-loop interaction. By integrating the most recent structural studies, the subtlety of each of these moves is brought to light.
Text kindly contributed by the authors.