According to the World Health Organization, antimicrobial resistance already kills 700,000 people each year and without government intervention, this number could rise to 10 million annually by 2050.
In this context, research and development of novel, next-generation antibiotics is urgently needed. To this end, finding new, effective targets is among the highest priorities. The advent of new technologies in RNA research have enabled the targeting of non-coding RNAs that play key roles in bacterial physiology, metabolism and pathogenesis.
T-box riboswitches are among such promising targets, as they control many essential genes in most Gram-positive bacteria, which include the largest group of clinically important human pathogens such as Clostridia, Staphylococci, Listeria, and Mycobacterium tuberculosis. As their name suggests, the T-boxes are genetic switches made of RNA, and interact with transfer RNAs (tRNAs).
tRNAs ferry amino acids, the building blocks of all proteins, to the ribosome to be assembled into proteins, a process that consumes significant resources. T-boxes are smart switches that can inspect tRNA couriers for their amino acid cargo, and then decide if the bacteria should commit scarce resources to synthesize or import amino acids.
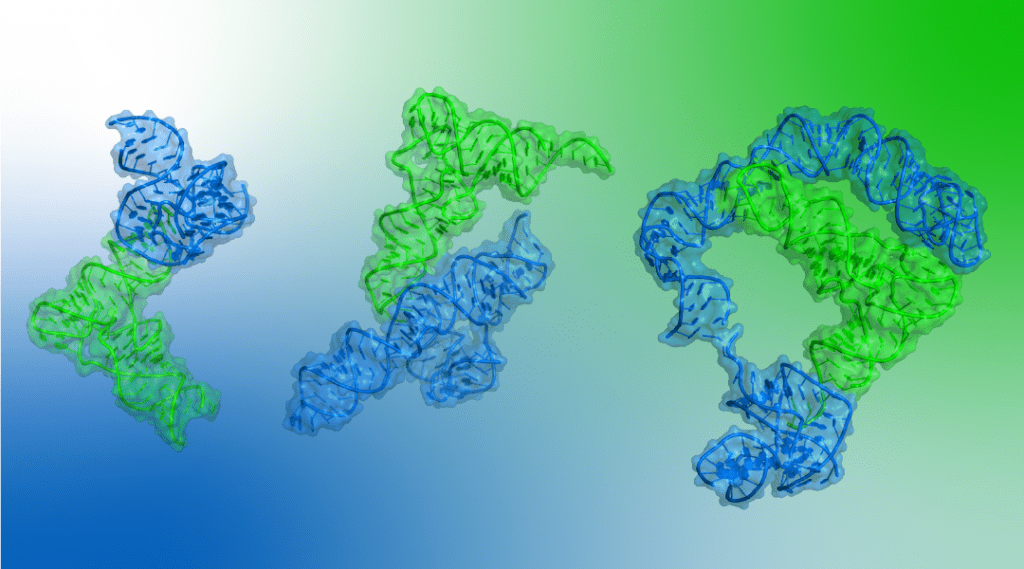
“If we understand how the T-boxes work, we could potentially design or identify drugs that block the bacteria’s ability to sense or respond to nutrient limitation — effectively starving themselves.” said Dr. Jinwei Zhang, an Earl Stadtman Investigator at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), part of the United States National Institutes of Health (NIH).
In a recent review published in WIREs RNA, Dr. Jinwei Zhang discusses work from his laboratory at the NIH which for the first time unraveled how the T-box riboswitches simultaneously recognize the three-dimensional structure of the tRNAs and sense the amino acids on them.
“Imagine one long RNA wraps around another on three sides, forming an RNA “sandwich” or ‘wrap’. This RNA sandwich gives the bacteria the ‘we need more food’ signal and directs the cell to make or gather more amino acids — the most important nutrients in the cell.” explains Dr. Shuang Li, a member of Dr. Zhang’s group. “It is remarkable that RNA alone can do all this complicated work without help from proteins.”
The RNA sandwich is constructed by a number of RNA-RNA contacts that work together. One particularly unusual contact involves a novel domain that acts as a bridge. “We found this bridging domain in a completely unexpected location and functioning in very surprising and clever ways. And yet in retrospect, it all makes sense, once you put two and two together.” commented Dr. Krishna Suddala, another member of Dr. Zhang’s group. “This domain, called “Stem II”, serves as a sandwich pick that reinforces the otherwise weak RNA-RNA interactions.”
These structures reveal how RNAs can engage each other in previously unknown ways using novel, highly defined interfaces. The existence of such conserved RNA-RNA interfaces makes it possible to target them with designer antibiotics.
“The unique advantage of the T-boxes as drug targets is that there are a multitude of similar instances of them controlling different essential genes.” Added Dr. Jinwei Zhang, “As a result, a single compound that effectively targets the shared T-box core could disable as many as ten metabolic pathways at the same time. This not only improves the efficacy of the antibiotic, but also dramatically reduces the occurrence of antibiotic resistance. Because it is much, much harder for the bacteria to come up with multiple drug-resistance mutations in all these different T-boxes at the same time.”
Reference: J. Zhang, Unboxing the T‐box riboswitches—A glimpse into multivalent and multimodal RNA–RNA interactions, WIRES RNA (2020), DOI: 10.1002/wrna.1600