Microscopic motors made of DNA are zooming in laboratories around the world. By tuning the make-up of these molecular machines, a group of researchers from Emory University have “turbo-charged” their speed and performance and are now rushing toward applications such as highly sensitive tests for respiratory viruses, like SARS-CoV-2 and RSV.
Nature has produced many molecular machines. For example, proteins, like myosin, act like motors that consume the cellular energy unit ATP to provide the mechanical work behind contracting muscles. Replicating this performance, however, has proven quite difficult for chemists, but researchers are now approaching the speeds of these natural machines.
The lab of Khalid Salaita, a professor of chemistry at Emory University and co-author of the paper describing the new turbo-charged DNA motors, wasn’t originally focused on this type of research. But casual discussions at a lab social event provided the spark. The group was pondering how interacting molecules like proteins, DNA and enzymes would behave if they were each fixed to different surfaces.
“We had this curiosity of what would happen if an enzyme was on a nanoparticle or microparticle, and the substrate, the thing the enzyme acts on, was also on another surface,” recalled Salaita. Since then, an entire branch of Salaita’s lab is now devoted to building synthetic machines made using the building blocks of the genetic code.
Powering motion with base pairing
The motors use short pieces of DNA or RNA made up of strings of base pairs connected to a chemical backbone. There are four types of bases and each one only binds to one other specific base, a phenomenon called complementary base pairing. This feature gives DNA and RNA it’s functionality and programmability.
For the motors, a spherical nanoparticle is coated in bits of DNA called the motors legs. The legs are positioned outward, so their bases are ready to bind to a complementary sequence. The motors begin to move when the legs bind to a strand of complementary RNA strand which acts like a track.
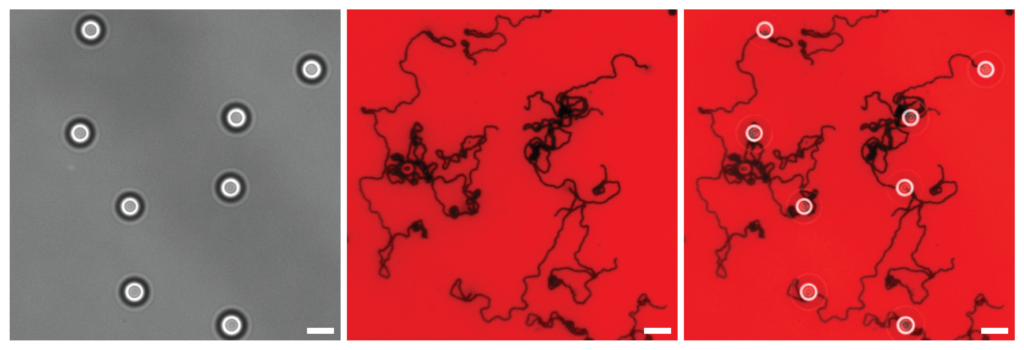
Movement continues when this bond is severed by an enzyme that specifically destroys RNA bound with a complementary strand. As the bound track is consumed, it releases the legs, freeing up others to bind with the next section powering the sphere along in a repeated cycle of binding and releasing.
Salaita’s team experimented with several designs to increase speed and processivity, meaning how long the motor will run. To turbo-charge performance, though, they had to overcome a fundamental trade-off between these to attributes.
Speed versus stability
The trade-off between speed and processivity arises because DNA motors tend to spontaneously release from the RNA track. A common solution has been to add more legs. According to Salaita, with more legs binding, the likelihood of spontaneous dissociation drops. However, this reduces speed because not all the legs bind and release in perfect coordination, leading to uneven motion and slower progress.
The spherical design helps because not all legs are engaged at once, but the team believed they could do more.
In the decade of researching these motors, there was one question the team hadn’t explored said Salaita. “What happens if you vary that DNA sequence?” he said. By experimenting with the ratios of the four possible bases used in the leg sequences, they found that legs made primarily of the bases adenine and thymine increased speed by three times without sacrificing processivity.
“We go to about 150 nanometers per second as our peak speed and that brings us in very close to the speed of biological motors like myosin,” said Salaita. This newfound speed allows the team to improve applications for the motor including highly sensitive viral detection.
Racing toward virus diagnostics
In 2023, the group uploaded a preprint paper to bioRxiv describing a proof-of-concept test for SARS-CoV-2. The tests use an RNA track complementary to both the motor legs and a target sequencing of viral RNA. If the motor encounters a section of track occupied by a piece of virus it will stop, triggering a positive result.
Modelling experiments predicted that a motor balancing high speed with low binding force could quickly scan lots of track and dissociate even if only a few molecules of virus bound to the track. In the current paper, the team used the new “turbo-charged” motors and found they could detect single nucleic acid targets, a vast improvement of sensitivity for this type of test.
The team continues to tinker with the motors and the viral tests. Due to the relatively low-cost and chip design, they believe the test could eventually screen for multiple viruses at once. Furthermore, the results are visible using only a smartphone.
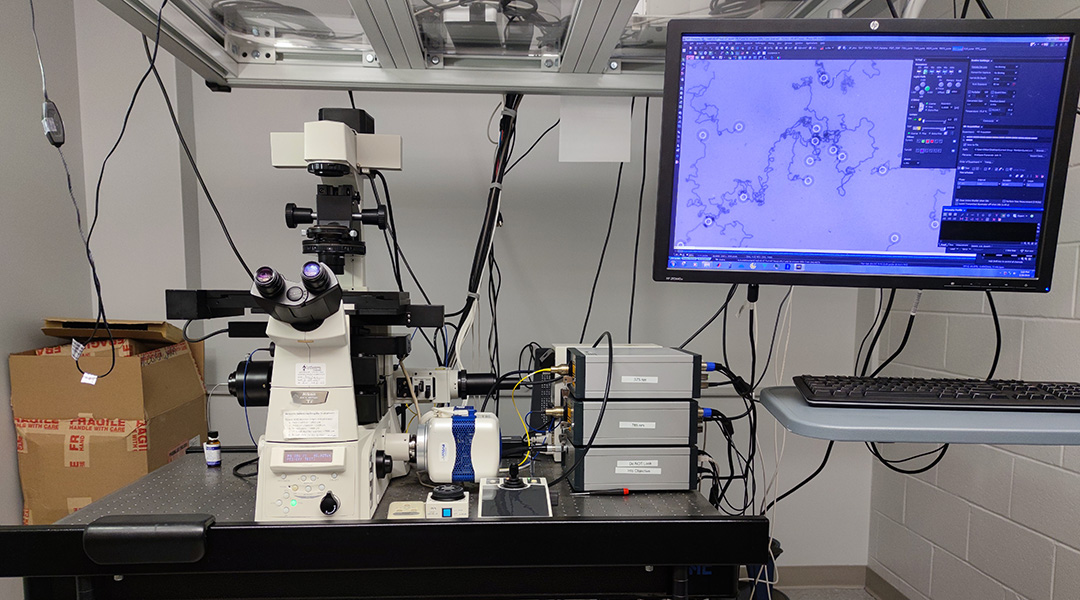
According to Salaita, the motors are big enough to be seen using a standard light microscope. “In fact, you can take a cell phone camera, put a plastic lens on it, just 10x magnification and detect the motion of the spheres,” said Salaita.
Reference: Khalid Salaita, et al., “Turbo-Charged” DNA Motors with Optimized Sequence Enable Single-Molecule Nucleic Acid Sensing, Angewandte Chemie International Edition (2024). DOI: 10.1002/anie.202316851
Feature image credit: Tharindu Rajasooriya