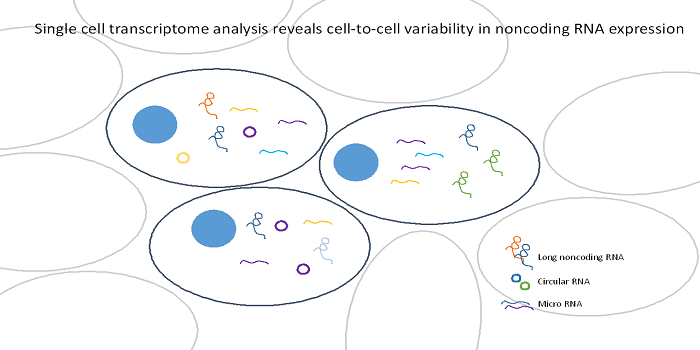
The human body is comprised of trillions of cells organized into distinct tissues. Until recently, it was thought that the single cells within a tissue were homogeneous, interchangeable building blocks. However, it is now possible to measure gene expression at the level of single cells. These data indicate that significant expression variability exists between cells, even in clonal cell lines. The biological role of this variation is still debated. One way to gain insight into the functionality of this heterogeneity is to examine single cell variation of non-protein coding RNA expression. Non-coding RNAs (ncRNAs) are highly involved in genomic regulatory function, and therefore evaluating ncRNA variation may yield insight into the potential role of single cell expression variation.
In a review published in WIREs RNA, a group survey the existing research on non-coding RNA expression at the level of single cells. Overall, the research suggests that non-coding RNAs show at least as much variability as protein-coding RNA, and some classes of non-coding RNA may display more single cell variability. The majority of studies focus on the expression profiles of long non-coding RNA (lncRNA), molecules of 200 base pairs or longer that play a key role in development and have been associated with a wide variety of human diseases (e.g., cancer, neurodegenerative diseases, cardiovascular diseases). Most of the studies report some level of cell-specific restriction of lncRNA expression, which may indicate that these molecules play important roles in differentiating cell lineages. There is also evidence that cell-specific expression patterns are especially pronounced for species-specific lncRNAs. These findings are in line with the idea that these cell-specific lncRNAs may have evolved to control species-specific cell fates.
Further research examining both single cell expression variation and functional outputs may yield a more detailed, quantitative picture of how expression variability impacts cell and tissue function. Developing additional technologies to measure a broad spectrum of non-coding RNAs and including their profile in single cell studies will be important for the Human Cell Atlas Project and other cell census studies.