Protein Science has published a Special Issue on Molecular Machines guest edited by Carlos Bustamante from the Laboratory of Single Molecule Biophysics at UC Berkeley.
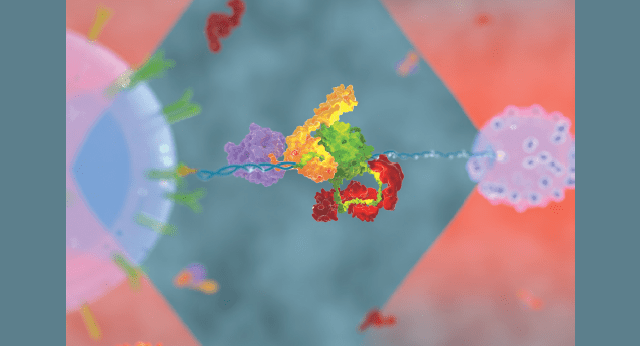
Each living cell, whether prokaryotic or eukaryotic, is a microscopic but complex structure. Its survival and wellbeing depend on replication, transcription, translation, splicing, protein degradation, energy generation, motility, and other cellular functions, all of which are performed by various proteins and enzymes. Carlos Bustamante describes them as “interconnected and highly coordinated protein machines”. In this Special Issue’s Guest Editorial, he writes, “These are assemblies of 5 or more polypeptide chains, contain various parts with specialized functions, and provide a localized environment where chemical species can interact and react in highly specific fashion. A protein machine is then a molecular “device” that, like its macroscopic counterpart, performs highly specialized functions requiring the conversion of chemical energy into mechanical work. This process—almost invariably—involves parts that move in some precise manner to produce forces, torques, and displacements.”
A substantial research effort focuses on the structure, function, thermodynamics, and mechanics of how these molecular machines operate inside the cell. The technological progress of the last two decades provided Structural Biology and Biophysics with high-tech imaging and fluorescence detection equipment, semi-automated and automated data and image acquisition and analysis software, single molecule methods of mechanical manipulation, and many others.
This Special Issue includes 1 Editorial, 9 Reviews and 7 Original Articles from several leaders of the field. They represent the best examples of research on single-molecule machines using optical tweezers, FRET, super-resolution imaging and other state-of-the-art structural biology and biophysical techniques.