The double‐stranded RNA‐binding protein family controls RNA editing, stability, and function in all eukaryotes.
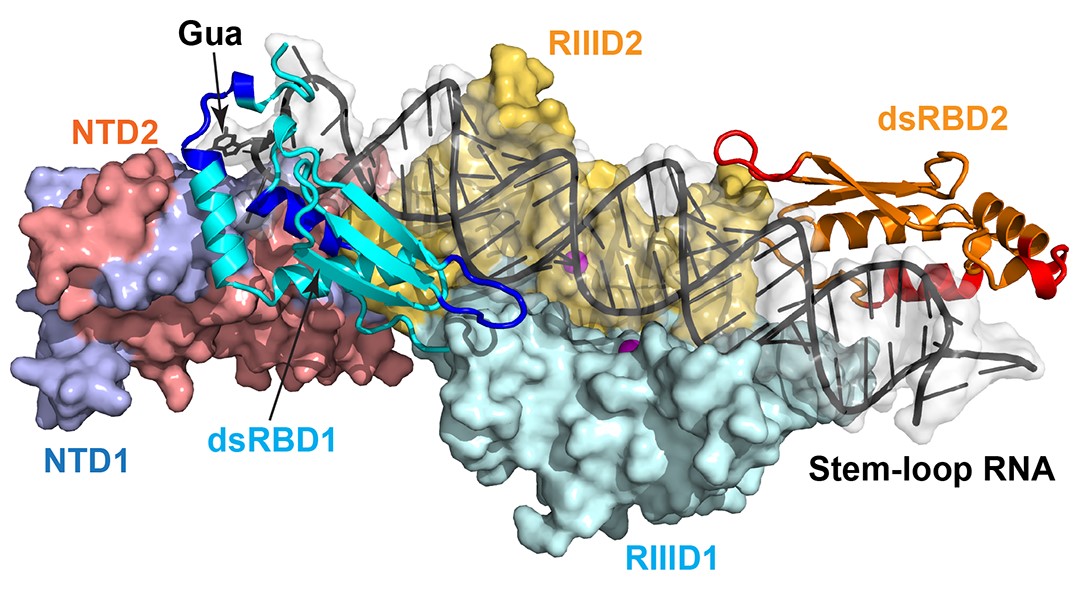

The double‐stranded RNA‐binding protein family controls RNA editing, stability, and function in all eukaryotes.
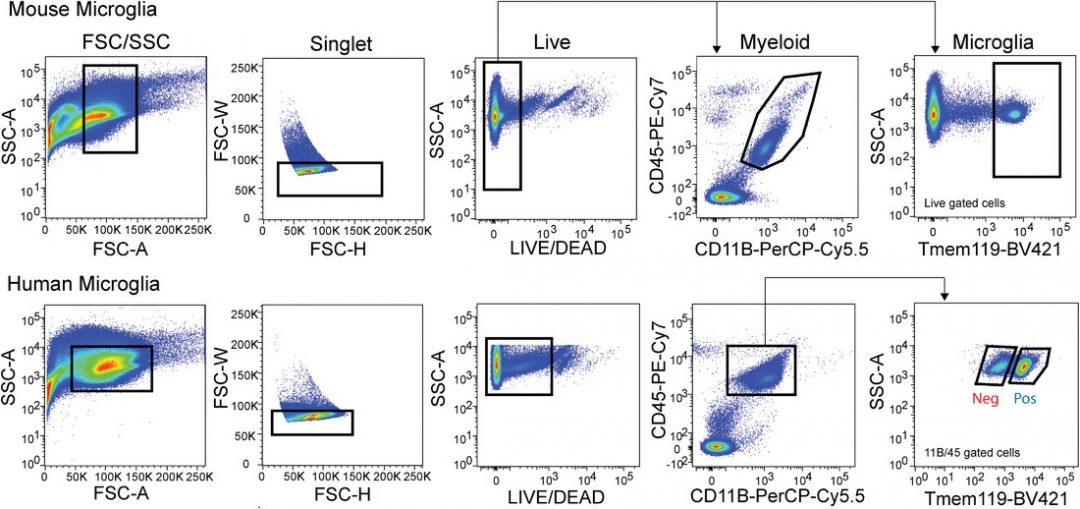
From gels to next‐generation sequencing – The evolution of RNA structural probing methods are discussed.

Mammalian DEAD‐box RNA helicase DDX5, its paralog DDX17, and their orthologs in Saccharomyces cerevisiae and Drosophila melanogaster define a subfamily of DEAD‐box proteins.
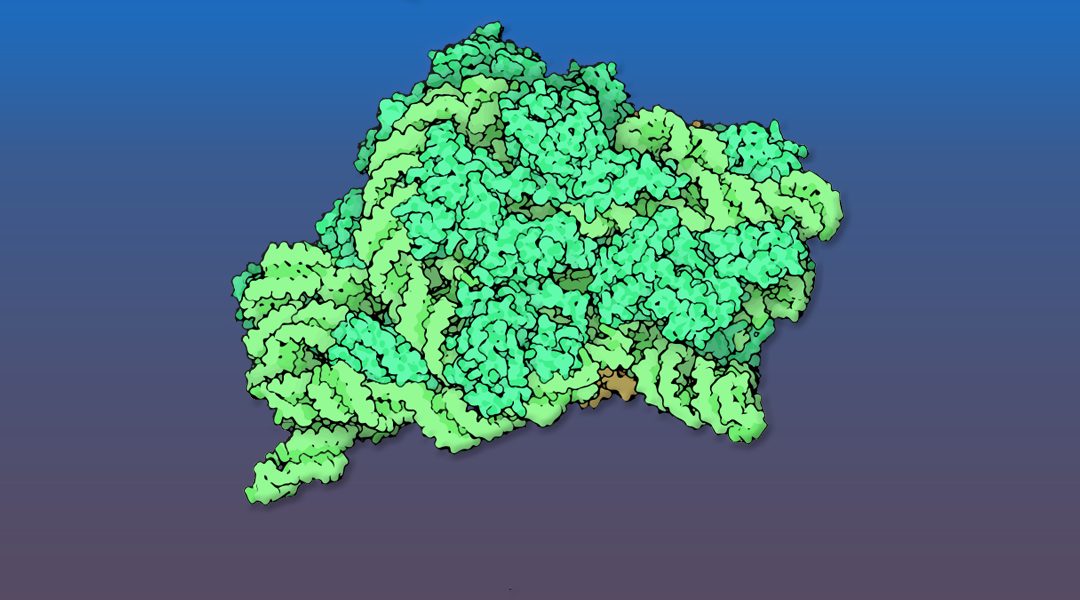
The intricate nuclear and cytoplasmic maturation steps of pre‐40S particles, the precursors to the small ribosomal subunits, in both yeast and human cells, are reviewed.
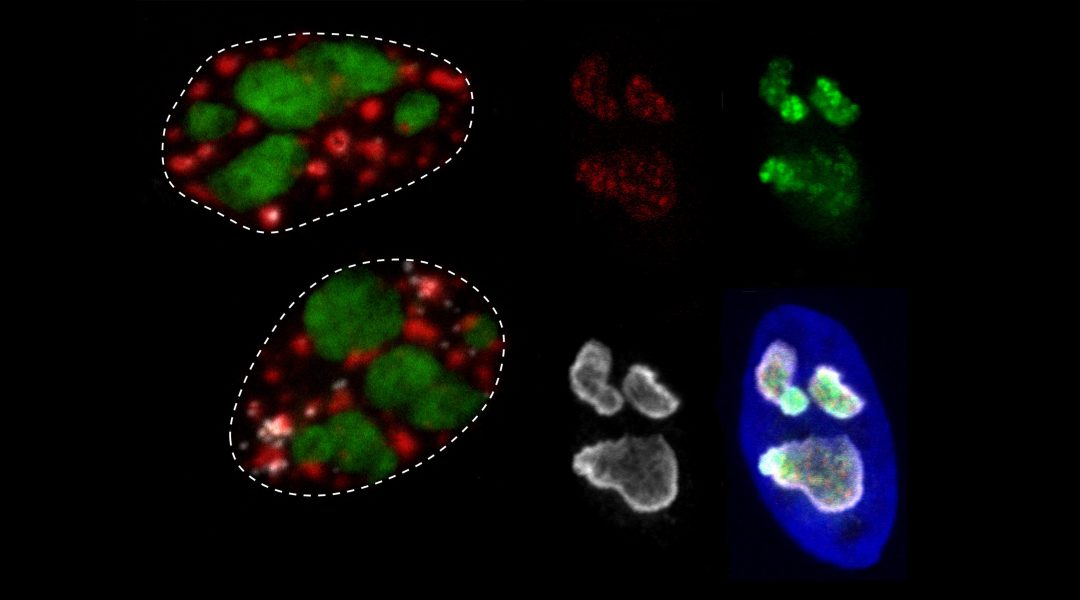
An analysis of nuclear body protein disorder that suggests MLO proteomes are significantly more disordered than structured cellular features.
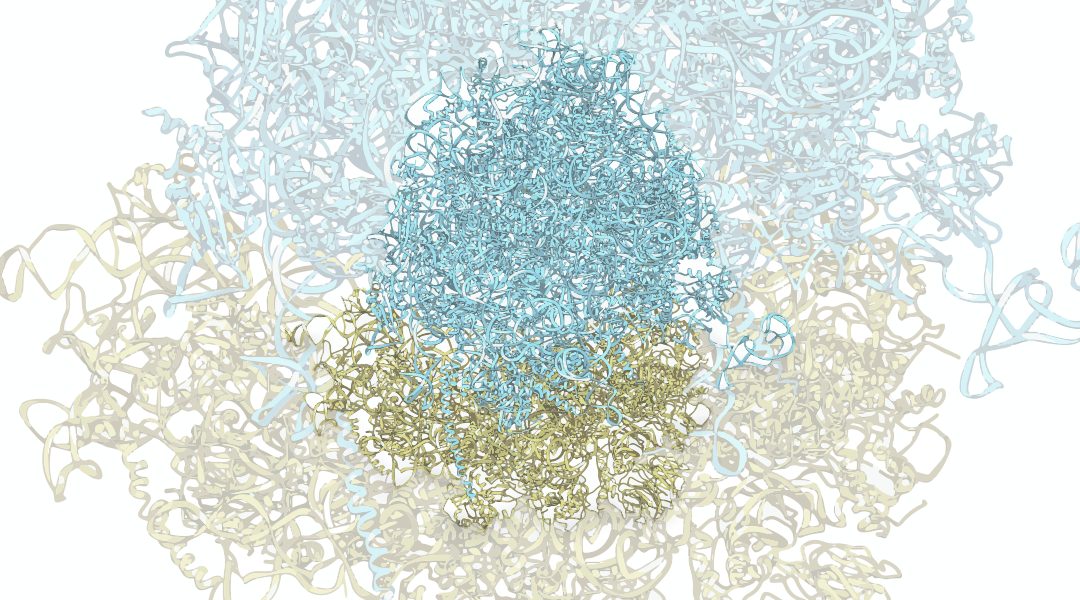
Poxviruses are an unusual family of large double‐stranded (ds) DNA viruses that exhibit an incredible degree of self‐sufficiency and complexity in their replication and immune evasion strategies.
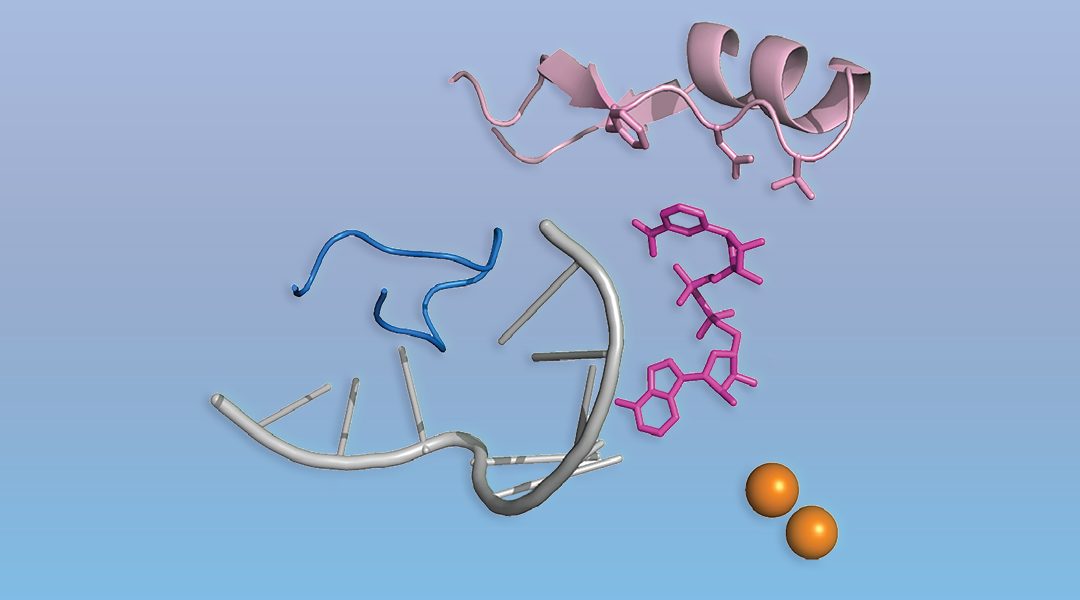
A new type of 5′‐RNA cap was discovered, and in contrast to the specialized eukaryotic m7G cap, the novel caps are abundant cellular cofactors like NAD+.
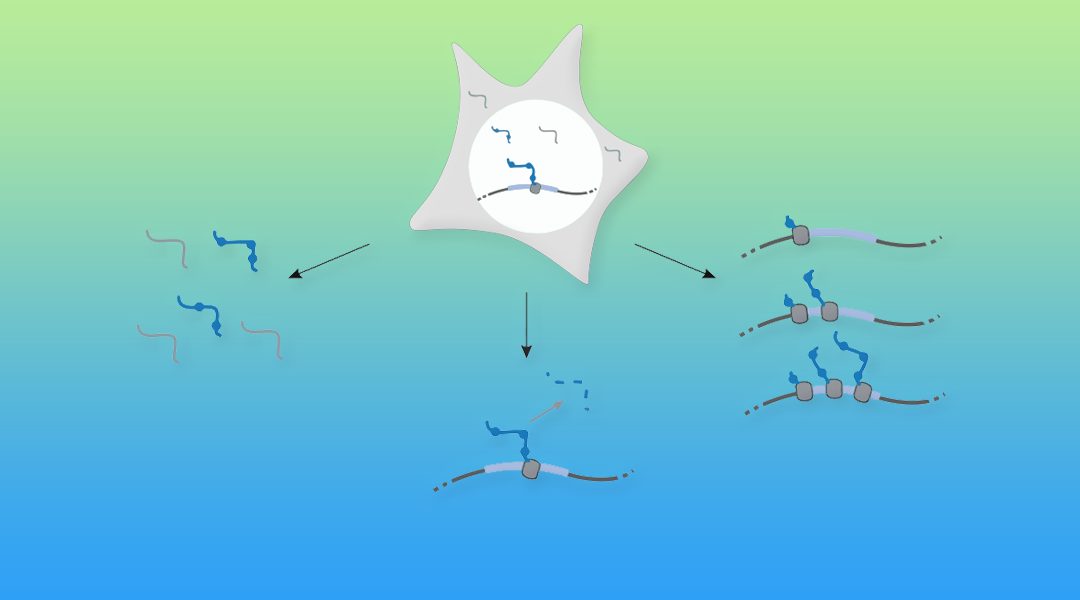
Cellular RNA levels are the result of a juggling act between RNA transcription, processing, and degradation. By tuning one or more of these parameters, cells can rapidly alter the available pool of transcripts in response to stimuli.
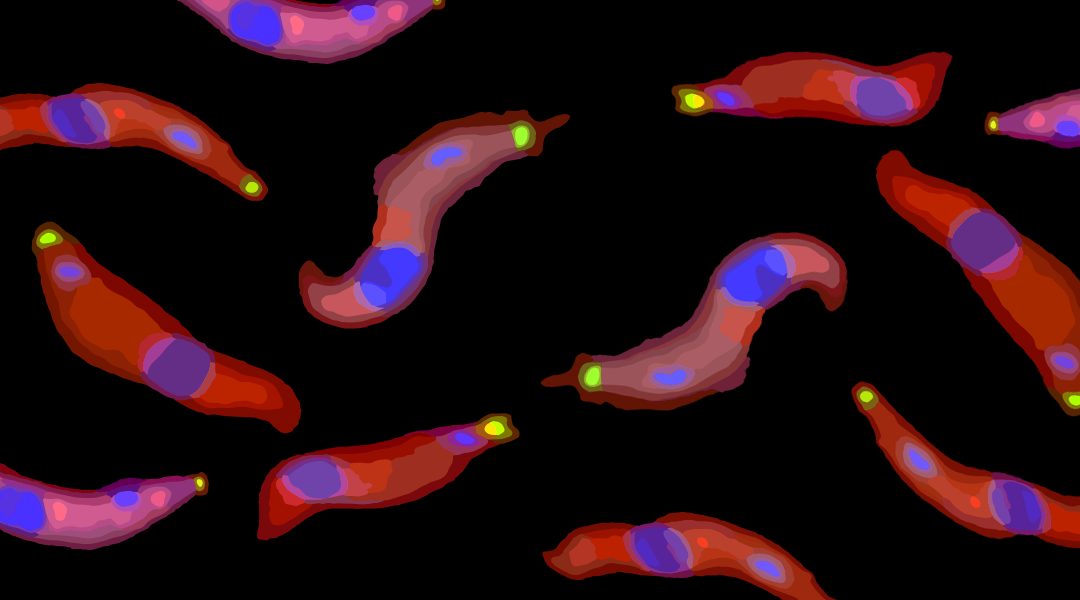
Single stranded RNAs with a free 5′ monophosphate end are susceptible to rapid degradation. Transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) are stabilized by hairpin structures and by “hiding” their 5′ ends within complex protein structures.
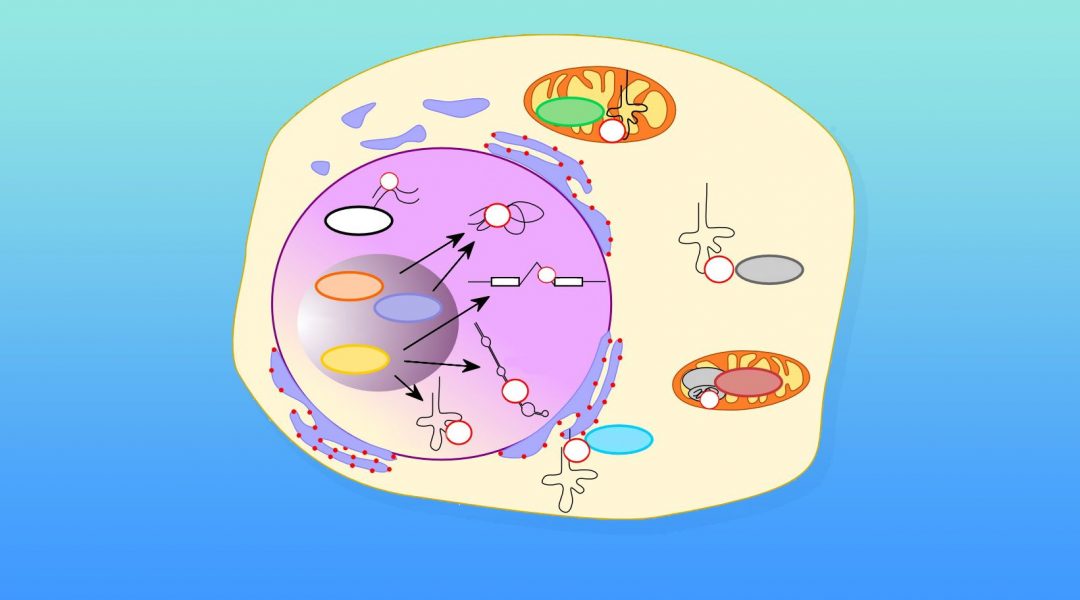
It is a well‐known fact that RNA is the target of a plethora of modifications which currently amount to over a hundred. The vast majority of these modifications was observed in the two most abundant classes of RNA, rRNA and tRNA. With the recent advance in mapping technologies, modifications have been discovered also in mRNA and in less abundant non‐coding RNA species.