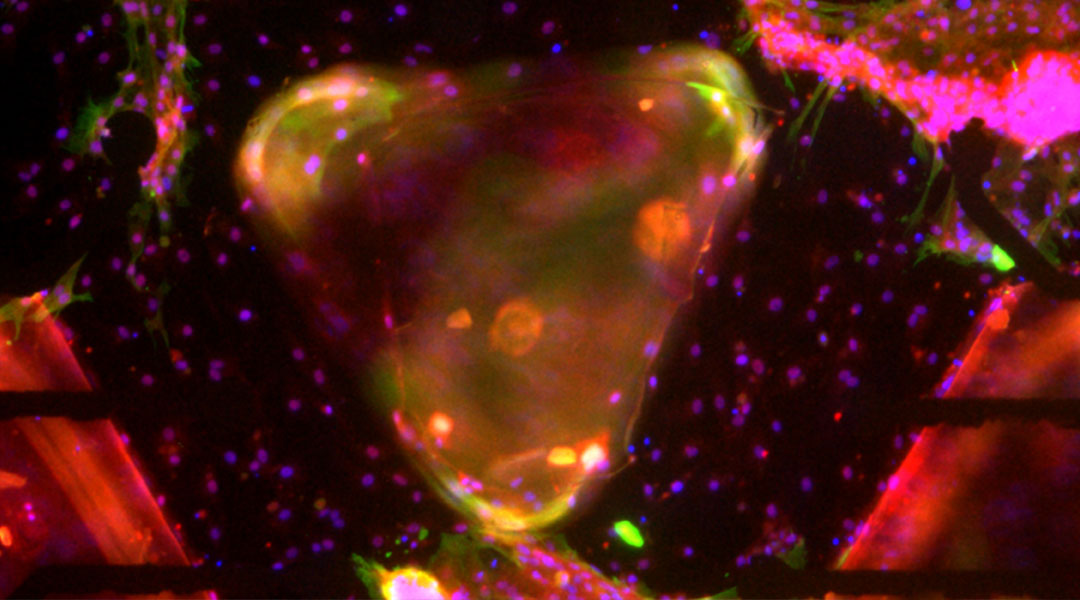
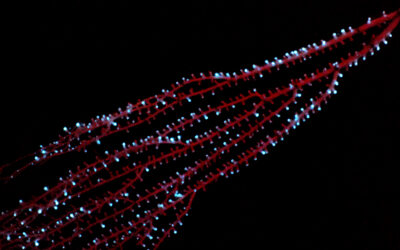
A 3-node bio-oscillator for solving graph coloring
Physical systems, including biological systems, have a natural ability to minimize their own energy to reach a ground state — a process that finds a natural analog to problems such as optimization and learning. Based on this ability, new computing paradigms can be developed with in the emerging field of biocomputing, which is an exciting new field aimed at creating computers from biological materials that mimic biological organisms or are used to study them.
In a recent study published in the journal Advanced Intelligent Systems, a biocomputing system constructed using heart muscle cells was shown to natively solve a prototypical constrained optimization problem e.g., graph coloring.
How do we build biocomputers?
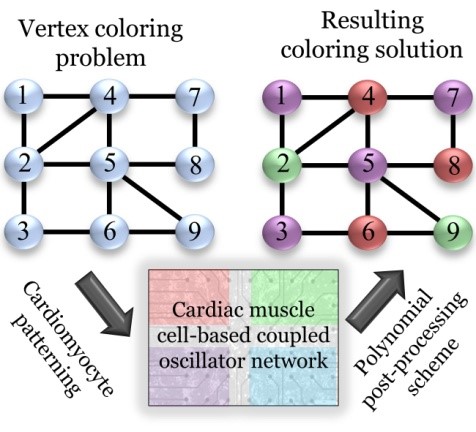
In such problems, a function is optimized using variables that are subject to constraints, such in the theoretical graph coloring problem where colors are assigned to the vertices of the graph such that no two vertices sharing the same edge have the same color. These types of problems have many real-world counterparts, such as in fault diagnosis, optimal path planning, scheduling, and resource allocation.
Current designs that explore biological components for computing leverage information processing units of cells, such as DNA replication, and gene and protein circuitry, to achieve simple computing tasks that often times relay on “Boolean logic” — a type of digital logic that operates on the idea that everything is 1 and 0 or true and false, and that forms the Boolean operators ‘OR’, ‘AND’, and ‘NOT’.
For instance, selective DNA duplication or manipulated gene expression using synthetic biology approaches can achieve some of the basic digital logic algorithm functions, such as ‘AND’ or ‘OR’ gates, within the living cell. For example, bacteria can be genetically engineered to have an ‘AND’ gate by inserting a certain readout gene with a detectable product (such as a florescent molecule) to be produced only if two input molecules are present in the environment at the same time.
While promising, these systems have limitations as certain classes of computational problems are fundamentally difficult to solve within the Boolean framework, and these cellular processes are inherently slow, requiring hours to days to complete a single computation.
The first biocomputers built using heart cells
Alternatively, electrically active cells — such as heart cells — that can function on the millisecond time scale are a potential alternative for developing faster biocomputers. Combined with collective computing algorithms where under certain conditions a collectively ordered state emerges from the complex dynamics of a non-linear system, networks of these types of cells in a synchronized state could transform biocomputing and lead to novel ways of storing and processing complex data.
To this end, muscle cells were first explored in 2017 by our group for information processing and were used to create the first functionally equivalent analogue of an electronic diode (i.e., a P-N junction, which is the basic computing function of a “one-way switch”) made entirely of living muscle-cells
Building on this earlier work, in the current study, the first ever cell-based computing fabric was created that could solve a 3-node graph coloring problem, demonstrating the potential of muscle-cell based circuits for performing complex computational tasks.
This new biocomputing unit is built using cardiac muscle cells, which act as natural electronic oscillators that emit periodic electronic signals. However, the key to their ability to form a functioning biocomputer is their ability to synchronize with one another when patterned and linked on a surface using microfabrication technologies.
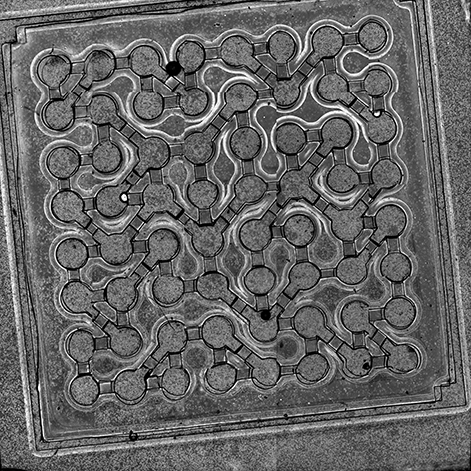
Initially, each cardiac muscle has its own beating frequency, but as they form connections predetermined by a designed pattern, the whole system reaches a stable beating frequency. In the body, this ensures that the heart beats at a stable pace. When linked in an artificial network that resemble integrated circuits used in everyday electronics, these cells also synchronize with one another and naturally converge to a minimum energy state.
This can be mapped to the “best solution” for a computationally hard problem such as graph coloring. The synchronized state emerges as an energetically favorable configuration instead of searching a huge solution space for every possible solution, which is what Boolean algorithms do. In other words, this behavior inherent to the cardiac muscle cells can be engineered to provide a biological way of finding a “best solution”, which is much faster than traditional Boolean-based computing.
In addition to their inherent connectivity, scalability, and parallel processing ability, the coupled bio-oscillator approach developed in this study requires minimal energy, mostly in the form of sugar, and as such can potentially be a low-power, sustainable alternative for current energy-demanding traditional computing approaches.
This study will open up a new field in which coupled networks of muscle cell-based oscillators are considered in new computing paradigms. In the long run, these results could lead to the development of faster and more efficient computing systems, as well as truly autonomous and unmanned vehicles that can make judgments and complex decisions beyond current abilities of electromechanical systems.
Such bio-based devices, computers, or vehicles, will have the ability to learn dynamically, self-diagnose, sense, and self-heal, while having low energy requirements. Such bio-based devices not only provide immediate interfacing ability with biological tissues, but also will have the advantage of collective analog computing that nature has evolved over billions of years.
Reference: Xiang Ren, et al., Cardiac Muscle Cell‐Based Coupled Oscillator Network for Collective Computing, Advanced Intelligent Systems (2020). DOI: 10.1002/aisy.202000253