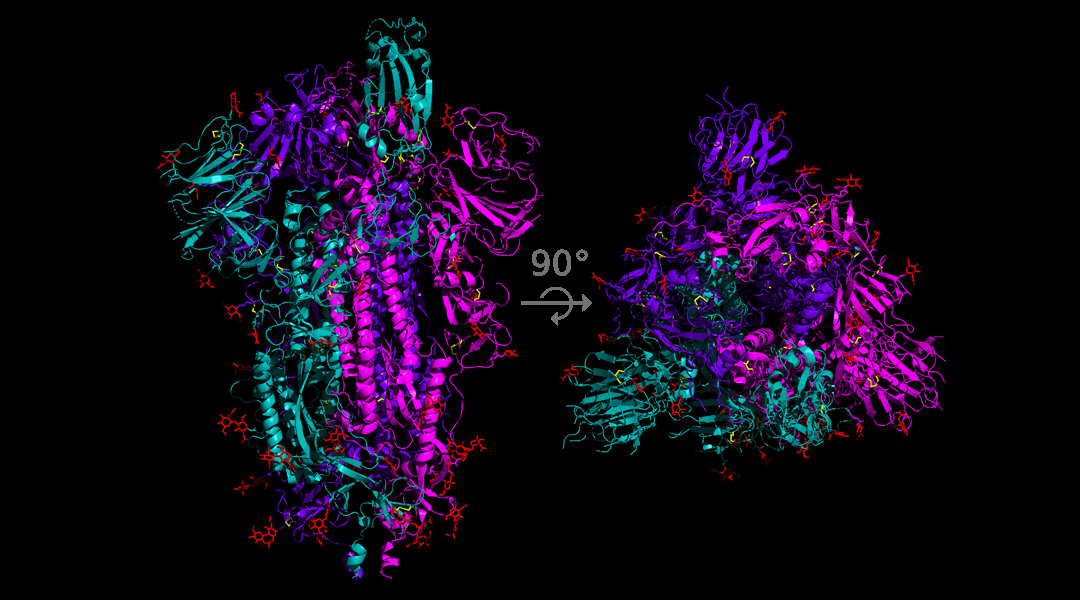As SARS-CoV-2 spreads around the world, its single strand of RNA is constantly mutating, and analysis of these mutations is helping governments and scientists around the world understand how the disease is spreading.
Mutations of the virus can be used in this way because every time the virus replicates, its copies contain the same mutations as the original. The available public data is being collected in an accessible manner by a group called nextstrain. The region-by-region phylogenies presented by this group show roughly the time and place that viral outbreaks originated.
One of the founding mutations in Europe, in late January, involved the infamous spike protein. The mutation was an A to G nucleotide swap, which caused the aspartate coded by this section of the RNA to be replaced by a glycine. From a functional point of view this is a bit like swapping the Lamborghini in your garage for a Nissan Micra. They both perform the basic function of ”car”, but the former has more bells and whistles.
This mutation is of particular concern to researchers who are working on developing a vaccine as many of the candidate vaccines under development rely on engineering an antibody response to the spike protein. If the variability in the spike protein structure can prevent the binding of antibodies then researchers might have to rethink this strategy.
An international team have recently published a pre-print aimed at building a tool to track all mutations of the spike protein in real time and have identified 14 mutations to the spike protein. For each of these mutations they conducted an analysis on the infectiousness of the resulting virus and what the mutation might mean for therapeutic research.
The main focus of the work was the A to G nucleotide swap, since data appears to show that when this mutant is introduced to an area it becomes the most common form of the virus. The data is not yet conclusive, and it is still possible that this mutated form is more common in the areas where it is found simply because it was the first version of the virus to arrive and has had more time to spread.
So while more study needs to be done to determine whether this mutant is more infectious, the authors of the pre-print offer some suggestions regarding possible mechanisms through which this mutation might confer selective advantage.
The first suggestion, is that the change to the protein structure caused by the aspartate/glycine swap might allow easier binding to human cells. The second suggestion is that this mutation could help neutralize immune responses to the virus.
Both of these propositions have yet to be thoroughly tested, the study simply flags these topics as areas for further study.
The authors do set out to test whether this mutant form increases the risk of hospitalization. To this end they followed the disease progression of 493 patients in England and found that there was no significant correlation between the mutation and likelihood of hospital admission.
This suggests that even if this version of the virus is more transmissible, it is not necessarily more dangerous.