According to the central dogma of biology, genetic information is stored in DNA and then passed on to messenger RNA, a single-strand of a gene serving as template for the production of proteins, the workhorses of the cell.
The importance of mRNA goes well beyond its role as a template, as evidenced in recent studies. Natural modifications of mRNA can regulate translation or alter stability, influencing the mRNA’s live cycle. Subcellular transport and localization of a distinct mRNA provide means for gene regulation with exquisite spatio-temporal control. However, to date the precise endogenous functions of these modifications and the dynamics of these processes is not fully possible, partly because the available methodology is still underdeveloped.
A versatile toolbox for the investigation of trafficking, localization, and interactions of RNA in living cells is required to study how RNA molecules orchestrate gene expression and understand malfunctions in disease.
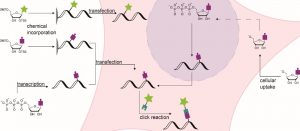
Schematic representation of various RNA labeling strategies.
A recent WIREs RNA review highlights covalent, state-of-the art RNA modification strategies and their diverse applications. These include visualization of a specific transcript or nascent RNA in living cells, as well as identification of internal modifications. The study brings together related concepts that have been used in different applications and thus, have not been gathered in one place before, providing critical analysis of the different approaches.
In cases of labeling, hijacking RNA-modifying enzymes for site-specific transfer of chemical handles which can subsequently be linked to a fluorophore has gained attention. Other covalent labeling strategies include incorporation of modified nucleotides instead of one of the four canonical building blocks of RNA, which can be used for visualization of a specific RNA.
These methods are not only applicable for labeling RNA but – with slight alterations of the involved chemistry – allow for the detection of small RNA modifications. If suitable nucleosides or their metabolic precursors are fed to cells, they are converted into the respective nucleotides and then statistically incorporated into all newly transcribed or modified RNA. This strategy allows for a temporal resolution of transcription.
This novel set of methods has the potential to significantly advance our understanding of the dynamics of RNA metabolism.