Many therapeutic agents are made of peptides and proteins. Over the last couple of decades, the application of protein-based drugs has been increasing due to their superiority over other types of drugs, such as their specificity and the variety of therapeutic roles which they could play. However, proteins are sensitive, having a fragile structure, which could be easily damaged during their delivery to the site of disease. On the other hand, proteins have large molecules which make it difficult for them to cross the cell membranes. In order to get the full therapeutic function from the protein-based drugs, they should be encapsulated in nano/micrometric carriers. These carriers can protect the protein molecules from damaging factors, such as degrading enzymes. On the other hand, these carriers can facilitate the cell internalization.
 DNA-based constructs have the potential to be functionalized for various applications, such as biosensing, biocatalysts, bioimaging, and the delivery of therapeutics. The feasible use of such constructs in biomedical applications has been inhibited by the possibility to produce and purify synthetic DNA at a large production scale and low cost. Rolling circle techniques, such as rolling circle amplification (RCA) and transcription, are a rapidly growing class of isothermal enzymatic reaction for nucleic acid amplification. These strategies, particularly RCA, provide a powerful alternative to produce large amounts of long DNA strands with desired sequences, further allowing the assembly of dynamic and complex DNA architectures.
DNA-based constructs have the potential to be functionalized for various applications, such as biosensing, biocatalysts, bioimaging, and the delivery of therapeutics. The feasible use of such constructs in biomedical applications has been inhibited by the possibility to produce and purify synthetic DNA at a large production scale and low cost. Rolling circle techniques, such as rolling circle amplification (RCA) and transcription, are a rapidly growing class of isothermal enzymatic reaction for nucleic acid amplification. These strategies, particularly RCA, provide a powerful alternative to produce large amounts of long DNA strands with desired sequences, further allowing the assembly of dynamic and complex DNA architectures.
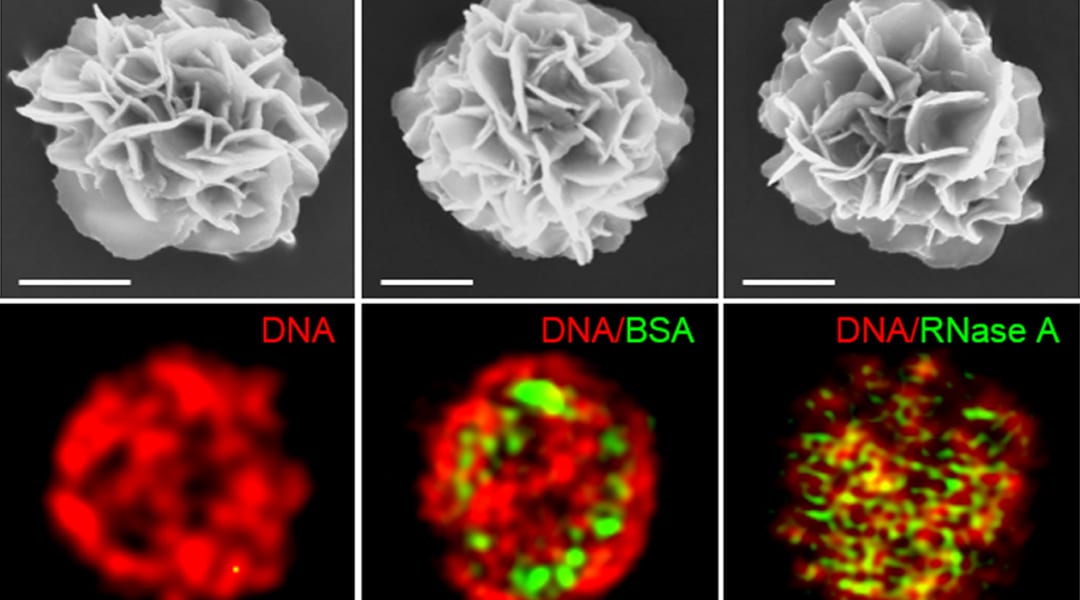
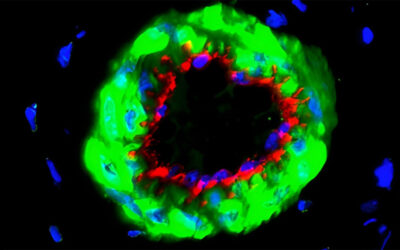
Researchers within Professor Molly Stevens’ group at Imperial College London describe the use of these techniques in their recent paper published in Advanced Materials. They show that a wide range of bioactive proteins, including enzymes, can be directly encapsulated in a three-dimensional DNA construct using the RCA technique. They produced protein-containing DNA particles by simply introducing protein solution in a typical RCA reaction, called “DNA flowers” due to their morphological features. The protein molecules are then simultaneously associated with the growing DNA strands and inorganic magnesium pyrophosphate crystals during the process, enabling the highly efficient entrapment of proteins.
This work suggests a simple, versatile, and new approach for fabricating a protein-DNA inorganic composite through a one-pot process. DNA flowers show good binding properties with specific biomolecules and high loading efficiency and enhanced stability of the protein cargoes.
Amongst a diverse set of biomolecules for encapsulation and inherent functionality and biocompatibility of the DNA component, the DNA flowers provide great opportunities to deliver therapeutic agents. Beyond protein delivery, this novel method will be applicable to a broad scope of applications such as biosensing, biocatalysis, and bioimaging.