New software and apps bring convenience to our daily life – and to drug discovery research too. Since most diseases are caused by malfunctioning proteins or DNA in the body, scientists strive to find the right molecule that binds to and interact with these biological targets, in order to rectify their physiological responses. First, scientists have to identify the likely locations on the target to which a drug molecule can bind. Second, a large number of potential drug molecules are investigated for their interactions with the target. Finally, scientists need to understand the impacts of a molecule on the target. Taken together, these tasks are exhaustive and challenging.
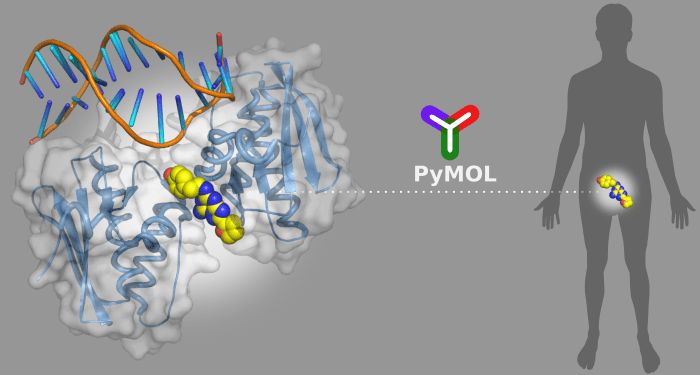
By visualizing the three-dimensional (3D) structures of the disease-related targets, scientists can better understand the underlying problems and design new molecular constructs to tackle the issues accordingly. PyMOL is a powerful tool to display the 3D structures of biological targets, and offers up to 12 different stereo visualization modes. Users can efficiently highlight and distinguish various important structural features in the targets, particularly the suitable binding sites for drug molecules.
Being a Python-based software, PyMOL has the advantage of embedding many useful Python-based plugins to enhance its functionalities for drug discovery calculations. Python itself is an “easy-to-script” programming language. Scientists without extensive programming experience can also develop plugins with great ease. Many available plugins online cover the key steps in computer-aided drug design, from simple tasks like in situ molecule editing to more complicated tasks like the analyses of drug-target interactions. Effectively, PyMOL has become a universal platform for drug design. In a recent review from WIREs Computational Molecular Science, Shuguang Yuan, H.C. Stephen Chan, and Zhenquan Hu provide an excellent introduction to present 3D structures visualization and computational drug design in PyMOL.