The study of virus-host interaction has been a long and meticulous feat aimed at deciphering the impact of viral infection, the response of host cell, and the complex interplay and cross-talk between the two. Upon infection, a tug of war is established between the virus and its host to either gain control of cellular functions, which are vital for viral replication, or mount an efficient antiviral response to prevent the infection from spreading. The result is either replication of the virus or control of the infection and clearance of the infected cell. Viruses have therefore evolved numerous ways to trick the cell and sabotage important defense pathways, allowing them to take control. These viral tricks are crucial in designing new antiviral drugs against viruses, such as Ebola and HIV.
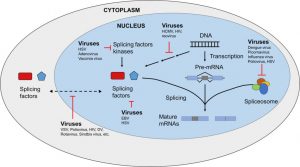
Insights into the modulation of cellular AS are triggered by viral infections.
As part of the maturation of messenger RNA, alternative splicing is an important step that allows parts of an RNA molecule to be removed or kept in the final transcript. Alternative splicing is widespread in higher eukaryotes, and it is estimated that nearly all genes in humans are alternatively spliced. It is a crucial phenomenon for the cell, as it enhances the diversity and function arising from genes and enables another layer of fine regulation for the activity of the proteins produced.
It has been known for a long time that some viruses, such as adenoviruses, human papillomaviruses, and HIV, usurp the cellular machinery involved in alternative splicing during infection to in turn splice their own genes. However, the impact of viruses on the cellular alternative splicing landscape was not studied thoroughly until very recently.
A WIREs RNA review highlights the recent research efforts to examine changes in alternative splicing during virus-host interactions, and their role on the interplay between viruses and host cells. A better understanding of how and why viruses modulate cellular alternative splicing will help to grasp the complete portrait of virus-host interaction. Elucidating the role of cellular alternative splicing in viral replication could improve the design of better oncolytic viruses, which are a promising treatment against cancer, and unveil potential new antiviral drugs exploiting this modulation.
Kindly contributed by the Authors.